













| General Information: |
|
| Name(s) found: |
suv3 /
SPAC637.11
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 647 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[ISS mitochondrion [IDA |
| Biological Process: |
mitochondrial RNA 3'-end processing
[IMP mitochondrion organization [RCA [NOT]RNA catabolic process [IMP |
| Molecular Function: |
ATP binding
[ISS RNA binding [IEA] ATP-dependent RNA helicase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFLETLIHGV TFCVPFFSKS ARIHTLSKDV FQQRKFPGNT LWAAALNRFT AYLFASKELS 60
61 SKQAYLAQDF VNVCKDASVF QNVYYYELKK NILTDLGFSD LKNSDKSLAL SKSSSTFDLQ 120
121 KIKKIHDCLL SEYRKYVRYQ ERIEETRPDL QKQLTDLKNP IEWYPGARKL RRHIIMHVGP 180
181 TNSGKTHRAL ERLKTCKKGI FAGPLRLLAH EIYNRLQANG IACNLYTGEE IRNDYPFPQV 240
241 VSCTVEMCNL STTFDVAVID EIQMMADPSR GYAWTQCLLG LQAKEIHLCG EESVVKLVRS 300
301 IAKMTQDDFT VYRYERLNPL HVAEKSLNGK LSELKDGDCV VAFSRKNIFT LKSKIDQALG 360
361 KKSAVIYGSL PPEVRNQQAS LFNSKSSDEN ILLASDAIGM GLNLGVKRIV FSDLKKFSGV 420
421 STIDIPVPQI KQIAGRAGRH NPNGSKQSAG IVTTLYQKDF AKLNRAMNLP TKNLFNACIG 480
481 AKDDLFFRYL SLFSDDIPQK LIFDRYFKLA KTTTPFVVSE GALSTFIIEY LDHIKGLTIK 540
541 DKIKLLGCPV LKHSKYAPLF IREIGCVIAQ GKRLQIYDLK SVPLEILERG IPTTETELQQ 600
601 LEQLHKLIVA YMWASIRYPA ILQNGAAEKT KAIAEAFLIK GISKLQK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
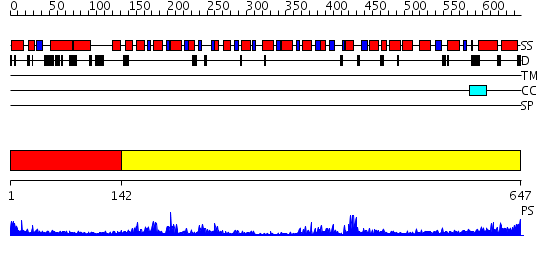
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..141] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [142..647] | 35.0 | Crystal structure of Escherichia coli transcription-repair coupling factor |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)