













| General Information: |
|
| Name(s) found: |
pmo25 /
SPAC1834.06c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 329 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cell division site [IDA actomyosin contractile ring [IDA medial cortex [IDA cell tip [IDA cytoplasm [IDA spindle pole body [IDA cytosol [IDA |
| Biological Process: |
regulation of cell shape
[IMP regulation of protein kinase activity [IMP regulation of cell separation during cytokinesis [IMP establishment or maintenance of cell polarity [IGI cell morphogenesis checkpoint [NAS unidimensional cell growth [IMP maintenance of cell polarity [TAS establishment of cell polarity [IMP regulation of bipolar cell growth [IMP actin filament reorganization involved in cell cycle [IMP establishment or maintenance of actin cytoskeleton polarity [TAS actin cortical patch localization [IMP cellular protein localization [IGI regulation of monopolar cell growth [IMP |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFLFNKRPK STQDVVRCLC DNLPKLEINN DKKKSFEEVS KCLQNLRVSL CGTAEVEPDA 60
61 DLVSDLSFQI YQSNLPFLLV RYLPKLEFES KKDTGLIFSA LLRRHVASRY PTVDYMLAHP 120
121 QIFPVLVSYY RYQEVAFTAG SILRECSRHE ALNEVLLNSR DFWTFFSLIQ ASSFDMASDA 180
181 FSTFKSILLN HKSQVAEFIS YHFDEFFKQY TVLLKSENYV TKRQSLKLLG EILLNRANRS 240
241 VMTRYISSAE NLKLMMILLR DKSKNIQFEA FHVFKLFVAN PEKSEEVIEI LRRNKSKLIS 300
301 YLSAFHTDRK NDEQFNDERA FVIKQIERL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
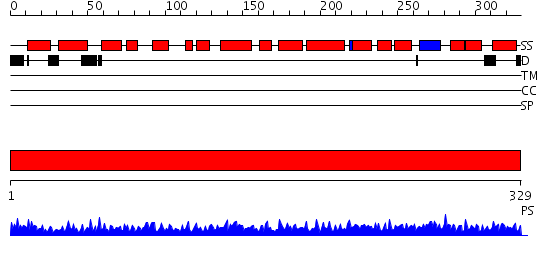
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..329] | 144.0 | Crystal structure of MO25 in complex with a C-terminal peptide of STRAD |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)