













| General Information: |
|
| Name(s) found: |
ubp13 /
SPAC22G7.04
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1115 amino acids |
Gene Ontology: |
|
| Cellular Component: |
PAN complex
[ISS cytoplasm [IDA |
| Biological Process: |
mRNA processing
[ISS postreplication repair [ISS ubiquitin-dependent protein catabolic process [IEA] |
| Molecular Function: |
poly(A)-specific ribonuclease activity
[ISS ubiquitin thiolesterase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDAWKEITKT TENDGVSTCS LSSIAFDPYS ELVWTGHKNG QIKSSFGPSL TSYTQFIGHE 60
61 GPVHQVLPQE RGVFSLSSKS LRLSNRKGTI RWRYHSVPCV QRSMIILYSC FLCSKHAVLI 120
121 SRDSDCIDYR AMFYQSRNNP EVVIGGYHQK LTVVNAERGI SIHKDKNVSD IFIMRRNRLL 180
181 CCGSTNGEII LRDPNSFQPV NKVVAHTGTI SDIDTSGNLL LSCGYSLRHG TYMLDPFVKV 240
241 WDLRNLSSLV PIPFPAGPTI IRMHPKLSTT AVVCSCSGQF HIVDTGNPLD AKLMQIPLTS 300
301 YLTGMDIAST GDAMVFTDVE DNIHLWSPLE NPSFSDLKLP IQLPNTSTET VQLENNDPLN 360
361 SIGLPYYKDE LLSSWSKYLI FDVGKPILDS NLLIAKQISE NSHPVPQEIK SFHRNQIIEV 420
421 PWLNRKLISE GATPKFHSER QKDIMSGNDI EGSASYFEEI EDTISGPDSI PKFYQRPVIK 480
481 YSKFGIEDFD FGFYNKTKYA GLETDITNSY CNSVLQLLSY VPSFSKAAIS HSLGPCDLME 540
541 CLLCELGFLF AMLKESTGRN CQATNFLRAF SNSSFAQSLG IVFDDYSDGT FPDSFVIQKF 600
601 TKFMLTEISR IADYEDKKDG TSFPVSFLLK SFCIPEMQTY RCGICGITSQ KIKSSLYIID 660
661 LHYPSQQLES ILSFEWLFKM SLDRRVDLPP GWCEYCLAHQ PFLLRSFIRS LPDCLFINTQ 720
721 VKHHEHWKLW ARKNWLPKKL HLRRVNDTMQ CVSQKISNLD KDQQSLSVYV LRGIIYEIRQ 780
781 NGEEPHFVST IRVSDNTSSD NPDDNRWYIF NDFLVKEVTE EEALTVHGPW KIPIIVYYEK 840
841 LDTKIPQWDE VSDYTLLYQP YSLNKNPPIN KIQPLTTDEM LYPKMLVGID SEFVALQQEE 900
901 TEVRSDGTKS TIKPSKLSLA RVSVLRGEGP NKGLPFIDDY VATDDKVTDY LTEYSGIHPG 960
961 DLDPDRSPYN VVPLKVAYKK LRLLVNAGCI FVGHGLQKDF RIINLLVPPE QVVDTVDLFF1020
1021 LSSRQRKLSL KFLAWYLLDE EIQLTEHDSI EDALTALKLY DCYDKLKSQG KLEETLDNIY1080
1081 EVGRRFKFRP PSVASMSLED RNSYGDESVI SNQTN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
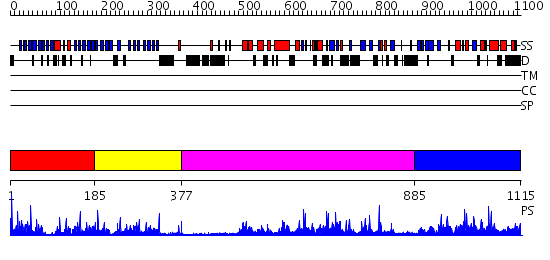
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..184] | 72.69897 | PAF-AH Holoenzyme: Lis1/Alfa2 | |
| 2 | View Details | [185..376] | 4.522879 | Crystal Structure Of Human Fibroblast Activation Protein alpha | |
| 3 | View Details | [377..884] | 60.0 | Crystal structure of HAUSP | |
| 4 | View Details | [885..1115] | 39.69897 | N-terminal exonuclease domain of the epsilon subunit of DNA polymerase III |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)