













| General Information: |
|
| Name(s) found: |
vma13 /
SPAC7D4.10
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 450 amino acids |
Gene Ontology: |
|
| Cellular Component: |
vacuolar proton-transporting V-type ATPase, V1 domain
[IEA]
vacuolar proton-transporting V-type ATPase complex [IC |
| Biological Process: |
ATP hydrolysis coupled proton transport
[IC vacuolar acidification [TAS ATP synthesis coupled proton transport [IEA] |
| Molecular Function: |
ATP binding
[IC proton-transporting ATPase activity, rotational mechanism [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNSDLELSN AASPPPVELD NSQVDEIINN VRCVAIPWQG YQRSGSLEEN ELQEIENLTG 60
61 KPLSAYVKTA EEDTTAYSNL FLKLLSMKDT PDVVNFALVK LADTLLNSNK FLSAFGPAFY 120
121 DFLEKDESYI NYLDDDSKLL FARVFALCSS SSPCSVAKAF TLFLEYLGKL MQSLNPLTRL 180
181 FAVQCLNGVL TLKAHRYALW AENTCSFRLA ELLRNSIGDT QLQYYSLFCF WQLTFESHIA 240
241 QDINKRFDLI KLLVQIIRSD TKTKVYRLVL AILVNLIDKA PKDTISTMLL EHVDKAVQLL 300
301 QKRKWADEDI TNYLDFITST LDESSKHLST FDMYKSELDT GILHWSPSHR SEDFWHQNAK 360
361 RLNEDNYALL KKLFHIVQYN EDNTSLAVAC HDLGAYIRSY PEGRSLIIKY GAKQRIMDLM 420
421 SHPDPEVRFE ALSTVQLLMT EVCFLSKITL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
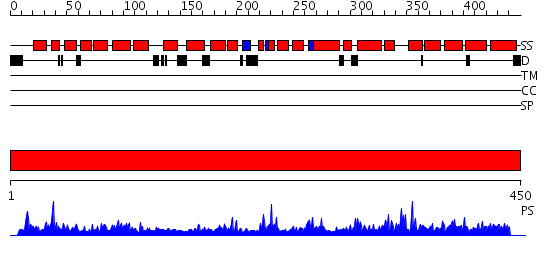
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..450] | 97.045757 | Regulatory subunit H of the V-type ATPase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)