













| General Information: |
|
| Name(s) found: |
tfb1 /
SPAC16E8.11c
[Sanger Pombe]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 477 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA cytosol [IDA holo TFIIH complex [ISS nucleotide-excision repair factor 3 complex [ISS |
| Biological Process: |
nucleotide-excision repair, DNA duplex unwinding
[ISS regulation of transcription [IC transcription initiation from RNA polymerase II promoter [ISS |
| Molecular Function: |
general RNA polymerase II transcription factor activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIRVFIVVKE GEDPTSLVFH FTGTPNAREN CDMITNELRN AIQRQREGSQ SKPQGANVDR 60
61 VNSTNLEKDI DLQESLLTNN PDLLQTFKEA VMKGHLSNEQ FWSTRLHLLR AHAVERSQQR 120
121 GPYNVLSTIK PKTVDNQMKV SLTGQQIHDM FEQHPLLRKV YDKHVPPLAE GEFWSRFFLS 180
181 KLCKKLRGDR ITPMDPSDDI MDKYLKDNEE VTKVSDEPIA SHLFDLEGND QNANVIAELR 240
241 PDITMRIDKE ALPFMKNINQ LSERLLEKSL GNSKRFNNEN EETYLKESGF HDLEEEASDS 300
301 KVVLKIKGQD QLLENNFVPS KNQVGLEPLP SLEALQHLYQ EDSISLNHIE HDSDSLAEAA 360
361 MQLTQSMREK HEFETHGTAS NLPIDIKNEI VLCHTVTIEF LHQFWNALLN TTFPDKKAMA 420
421 PLQNALLNSK KRVEAIASQA QEQNVDPKLV YELVSCTLTS IDTSISEYQR RMSYPPA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
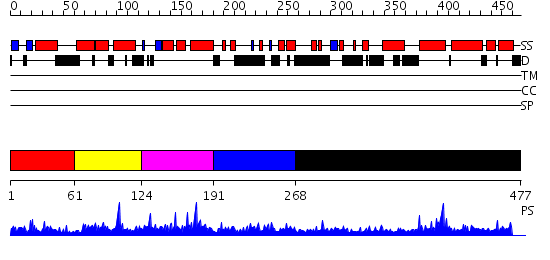
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..60] | 1.035993 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [61..123] | 12.154902 | No description for 2diiA was found. | |
| 3 | View Details | [124..190] | 1.74 | Solution structure of the BSD domain of human Synapse associated protein 1 | |
| 4 | View Details | [191..267] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [268..477] | 2.036983 | View MSA. Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)