













| General Information: |
|
| Name(s) found: |
thp1 /
SPCC965.05c
[Sanger Pombe]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 325 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
DNA repair
[IC mismatch repair [IEA] |
| Molecular Function: |
uracil DNA N-glycosylase activity
[IDA mismatched DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNDIETRDTG TKNDNSSEFN LSVKSHKRKR SFDDENLELE ESREETSGGI LKKAKTQSFS 60
61 ESLERFRFAH AGSNNEYRKT DVVKNSDTDN GLLKSAVETI TLENGLRNRR VNVTKKSTLK 120
121 ASVKKSTLKK KNEVDPALLQ GVPDYICENP YAIIVGLNPG ITSSLKGHAF ASPSNRFWKM 180
181 LNKSKLLEGN AEFTYLNDKD LPAHGLGITN LCARPSSSGA DLRKEEMQDG ARILYEKVKR 240
241 YRPQVGLFIS GKGIWEEMYK MLTGKKLPKT FVFGWQPEKF GDANVFVGIS SSGRAAGYSD 300
301 EKKQNLWNLF AEEVNRHREI VKHAV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
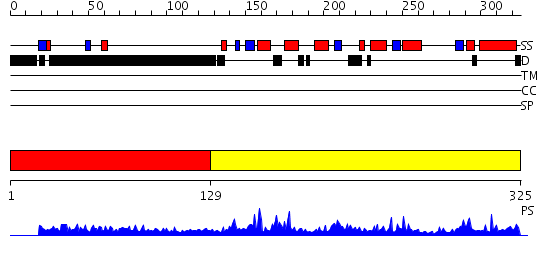
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..128] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [129..325] | 59.0 | Crystal Structure of SUMO1-conjugated thymine DNA glycosylase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)