













| General Information: |
|
| Name(s) found: |
spo6 /
SPBC1778.04
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 474 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA nucleoplasm [ISS |
| Biological Process: |
ascospore formation
[IMP signal transduction [IC] regulation of catalytic activity [IC meiosis II [IMP protein amino acid phosphorylation [ISS regulation of phosphorylation [IC |
| Molecular Function: |
zinc ion binding
[IEA]
protein kinase regulator activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDFYSVKSQP FVRSPLVDQN PSIQNINEEV KRDIQNPLSY KTETSDKELC QTAACATSCS 60
61 DWYPQQQTHM PHQNAFDSAK ATAKMALPPT AFSNYCVKPS LTRNKDIPRT SIRVSKLRYW 120
121 QRDYRLAFPN FIFYFDNVDE EIKRRVTQKI NNLGAKVATL FTFEVTHFIT TRTTDPEMCQ 180
181 PNDVLYLSKT ANMKIWLLDK LLNRILFTLL NSDSLVNTSA SCLQSLLDGE KVYGTSDKDF 240
241 YVPSKNVEYF REYFLCIRDL SQYYKPIAVR EWEKTLDSGE ILWPSLAITA QGRCPFNTGR 300
301 RRELKITKHN HPAHEIRKQL LSCTNQTNQN NVVKNSASVL VRQIMGDYNI TESAVDGAKQ 360
361 MPTEFPKPEN LLPVEKRAAM SPLNLLEPRL INKQNTLANQ SPRQPPNAFD ADPLAHKKVK 420
421 IETKSGYCEN CCERYKDLER HLGGKHHRRF AEKDENFQGL DDLFLLIRRP IRTN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
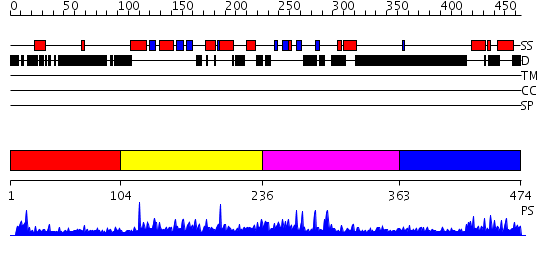
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..103] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [104..235] | 1.53 | The third BRCA1 C-terminus (BRCT) domain of Similar to S.pombe rad4+/cut5+ product | |
| 3 | View Details | [236..362] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [363..474] | 6.028996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)