













| General Information: |
|
| Name(s) found: |
rsv1 /
SPBP4H10.09
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 428 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA spindle [IDA |
| Biological Process: |
negative regulation of cellular carbohydrate metabolic process by repression of transcription
[IEP negative regulation of carbohydrate metabolic process [IEP gene-specific transcription from RNA polymerase II promoter [RCA cellular response to glucose starvation [IMP negative regulation of transcription from RNA polymerase II promoter, meiotic [IEP |
| Molecular Function: |
transcription factor activity
[NAS zinc ion binding [IEA] specific RNA polymerase II transcription factor activity [RCA specific transcriptional repressor activity [IEP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKSYECPFCK RVFHRQEHQV RHIRSHTGEK PFECSYPSCK KRFTRRDELI RHVRTHLRKA 60
61 LVTPEQTLDV NLHKAPDSKP EGDKSTGQEA DKSQNQSKDG SITDPVQAAV LALSVAYAKP 120
121 TSVSLSPTDL QAQSKLIEKP RRRSASNATG SLNKKNQDPL RRFSISESAG AAAPTPSPSN 180
181 SKSPPSENRK NRLQNVLSPI ASNNVPDFNQ NYPTESNPMF LSTPRFQNTN GQRTLTVPVS 240
241 VWDARQPPTS SRSGLPLSVM YNHFPSVPIP PATVNDTSME YYLPNAYPHP TGISLPFYPF 300
301 DSGIPVSPNI PVSPSSSFVP MYPTTFPSSK PQIVNAPPAP SYFSPGSSFG AQLCANGSTL 360
361 LRADTKQYGL ALPKITNSNL ISPNQTFNPV KSSVKALPTL EPPSSPSHAT ATSSLHTLFH 420
421 TAPSRSYD |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
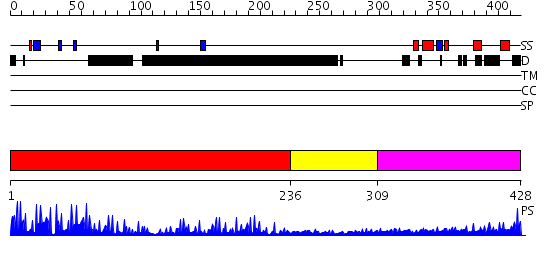
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..235] | 23.39794 | Transcription factor IIIA, TFIIIA | |
| 2 | View Details | [236..308] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [309..428] | 1.01 | No description for 2j63A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)