













| General Information: |
|
| Name(s) found: |
rpn7 /
SPBC582.07c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 409 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA proteasome regulatory particle, base subcomplex [ISS cytosol [IDA proteasome complex [IDA nuclear proteasome complex [IDA |
| Biological Process: |
proteasomal ubiquitin-dependent protein catabolic process
[IMP mitotic sister chromatid segregation [IMP proteasome localization [IGI regulation of cell cycle [RCA proteasome assembly [IGI mitotic cell cycle [IMP ubiquitin-dependent protein catabolic process [ISS |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MCSPSATLTH RTMTEKARTV SDLTISQAIF ELSSPFLENK SQKALDTLFS AIRDHDLAPL 60
61 YKYLSENPKT SASIDFDSNF LNSMIKKNEE KLAEFDKAIE DAQELNGEHE ILEAMKNKAD 120
121 YYTNICDRER GVQLCDETFE RATLTGMKID VLFSKIRLAY VYADMRVVGQ LLEKLKPLIE 180
181 KGGDWERKNR LKAYQGIYLM SIRNFSGAAD LLLDCMSTFS STELLPYYDV VRYAVISGAI 240
241 SLDRVDVKTK IVDSPEVLAV LPQNESMSSL EACINSLYLC DYSGFFRTLA DVEVNHLKCD 300
301 QFLVAHYRYY VREMRRRAYA QLLESYRALS IDSMAASFGV SVDYIDRDLA SFIPDNKLNC 360
361 VIDRVNGVVF TNRPDEKNRQ YQEVVKQGDV LLNKLQKYQA TVMRGAFKV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
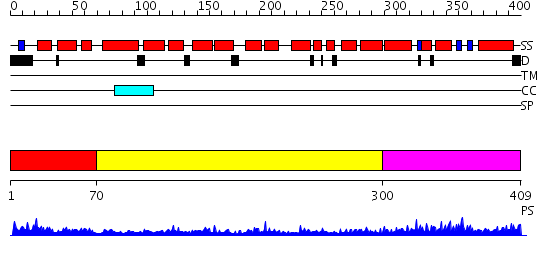
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..69] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [70..299] | 5.522879 | Crystal structure of an 8 repeat consensus TPR superhelix (trigonal crystal form) | |
| 3 | View Details | [300..409] | 11.69897 | Solution structure of the PCI domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)