













| General Information: |
|
| Name(s) found: |
rad16 /
SPCC970.01
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 892 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IC nucleotide-excision repair complex [ISS spindle pole body [IDA |
| Biological Process: |
nucleotide-excision repair, DNA incision, 5'-to lesion
[TAS meiotic gene conversion [IMP nucleotide-excision repair [IGI gene conversion at mating-type locus [IMP |
| Molecular Function: |
DNA binding
[ISS protein heterodimerization activity [IPI single-stranded DNA specific endodeoxyribonuclease activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 METKVHLPLA YQQQVFNELI EEDGLCVIAP GLSLLQIAAN VLSYFAVPGS LLLLVGANVD 60
61 DIELIQHEME SHLEKKLITV NTETMSVDKR EKSYLEGGIF AITSRILVMD LLTKIIPTEK 120
121 ITGIVLLHAD RVVSTGTVAF IMRLYRETNK TGFIKAFSDD PEQFLMGINA LSHCLRCLFL 180
181 RHVFIYPRFH VVVAESLEKS PANVVELNVN LSDSQKTIQS CLLTCIESTM RELRRLNSAY 240
241 LDMEDWNIES ALHRSFDVIV RRQLDSVWHR VSPKTKQLVG DLSTLKFLLS ALVCYDCVSF 300
301 LKLLDTLVLS VNVSSYPSNA QPSPWLMLDA ANKMIRVARD RVYKESEGPN MDAIPILEEQ 360
361 PKWSVLQDVL NEVCHETMLA DTDAETSNNS IMIMCADERT CLQLRDYLST VTYDNKDSLK 420
421 NMNSKLVDYF QWREQYRKMS KSIKKPEPSK EREASNTTSR KGVPPSKRRR VRGGNNATSR 480
481 TTSDNTDAND SFSRDLRLEK ILLSHLSKRY EPEVGNDAFE VIDDFNSIYI YSYNGERDEL 540
541 VLNNLRPRYV IMFDSDPNFI RRVEVYKATY PKRSLRVYFM YYGGSIEEQK YLFSVRREKD 600
601 SFSRLIKERS NMAIVLTADS ERFESQESKF LRNVNTRIAG GGQLSITNEK PRVRSLYLMF 660
661 ICIKTLKVIV DLREFRSSLP SILHGNNFSV IPCQLLVGDY ILSPKICVER KSIRDLIQSL 720
721 SNGRLYSQCE AMTEYYEIPV LLIEFEQHQS FTSPPFSDLS SEIGKNDVQS KLVLLTLSFP 780
781 NLRIVWSSSA YVTSIIFQDL KAMEQEPDPA SAASIGLEAG QDSTNTYNQA PLDLLMGLPY 840
841 ITMKNYRNVF YGGVKDIQEA SETSERKWSE LIGPEAGRRL YSFFRKQLKD YE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
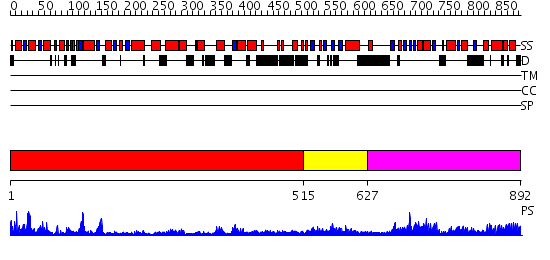
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..514] | 56.39794 | Crystal structure of Pyrococcus furiosus Hef helicase domain | |
| 2 | View Details | [515..626] | 2.64 | Crystal structure of Pyrococcus furiosus Hef helicase domain | |
| 3 | View Details | [627..892] | 39.0 | XPF from Aeropyrum pernix, complex with DNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.76 |
Source: Reynolds et al. (2008)