













| General Information: |
|
| Name(s) found: |
nup61 /
SPCC18B5.07c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 565 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear membrane [IDA nuclear pore [TAS |
| Biological Process: |
mitotic cell cycle spindle assembly checkpoint
[IMP nucleocytoplasmic transport [TAS |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKRGADHQL TKDQDDSDDD RHGPVEVPKE ASADVMATRK IAKPKSRKRP TSGVSSPGIF 60
61 ANLAAKPVSL PASTTQFTFG KPAVTANNDS DIHLKKRGLN KSFIDAVIKS VDNNPFGNLS 120
121 PLFDEYRQHF SSIEKKPAEG NAFIVSTSFL SNVFLEQPTS NAVVSEVNPQ QQKSQDSSSF 180
181 VTEKPASSEK EDKEKPLVPP GAPRFGFSAP ALGSSFQFNS SAFTPKGSFG EKSATEAEAK 240
241 EKETSSNQTA TGTAATTTNQ FSFNTAANPF AFAKKENEES KPLTPVFSFS TTMASADASK 300
301 ETKQTHETKD SKSEESKPSN NEKSENAVEP AKGNTMSFSW TPDKPIKFDT PEKKFTFTNP 360
361 LSSKKLPASS DVKPPSAAAV GFSFGTTTNP FSFAAPKSSF PTSSTPASVG AEKSEETSNG 420
421 NKSEQEEKEN GNDETRSNDS LVSGKGKGEE NEDSVFETRA KIYRFDATSK SYSDIGIGPL 480
481 KINVDRDTGS ARILARVEGS GKLLLNVRLC QDFEYSLAGK KDVKVPAAST DGKSIEMYLI 540
541 RVKEPSTAEK LLAELNEKKV SKSEN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
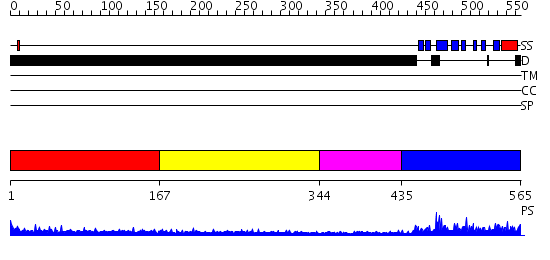
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..166] | 5.176995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [167..343] | 2.180994 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [344..434] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [435..565] | 42.0 | Nuclear pore complex protein Nup358 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)