













| General Information: |
|
| Name(s) found: |
mcs4 /
SPBC887.10
[Sanger Pombe]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 522 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell division site
[IDA cytoplasm [IDA |
| Biological Process: |
stress-activated protein kinase signaling pathway
[IGI response to arsenic [IMP two-component signal transduction system (phosphorelay) [IEA] regulation of two-component signal transduction system (phosphorelay) [IC] cellular response to oxidative stress [IC regulation of transcription, DNA-dependent [IEA] response to hydrogen peroxide [IMP regulation of mitotic cell cycle [IMP |
| Molecular Function: |
protein binding
[ISS]
two-component response regulator activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRIWFKKVPD GITSSVILSE DHLVDDLKDA IARKFPIRIS QYYDAPELSI RVVAPPNASS 60
61 ELQSRELSPN ESILFVMETY YPHGQDFNDA LLVASPDTSV ALRYRSSQLS SSTFESTPPV 120
121 FSEYPPNIIP TPANETVPRI KQPSIALDSL ESPVSAPSRH QSTYSYKGGP LNYNLRNASR 180
181 TRSHQTLPSS NVNKTGVLLL PRSSRQQTLA SRPSLPDLTS ADKSQPSDEA ESITRKNSIG 240
241 MSTRSDESTA EKLAKAEVAT PTNSRSISHS SLYTKQSGTA GVLPAVNADI DAANRMNPDI 300
301 SSQFPIADNK DPLNADTQAH LGFPSNQIDG IVGTSPVNVL TSPGIGAKAP FASLLEGVIP 360
361 PINVLIVEDN IINQKILETF MKKRNISSEV AKDGLEALEK WKKKSFHLIL MDIQLPTMSG 420
421 IEVTQEIRRL ERLNAIGVGA PKLTQPIPEK DQLNENKFQS PVIIVALTAS SLMADRNEAL 480
481 AAGCNDFLTK PVSLVWLEKK ITEWGCMQAL IDWNGWCRFR GR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
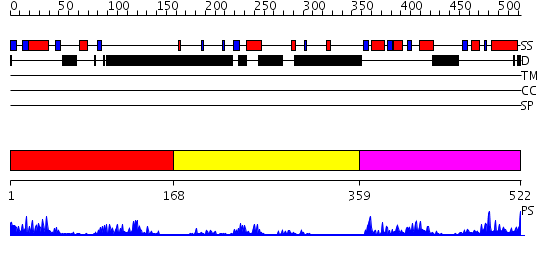
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..167] | 18.39794 | Structure of the entire cytoplasmic portion of a sensor histidine kinase protein | |
| 2 | View Details | [168..358] | 4.69897 | Histidine kinase PhoQ domain | |
| 3 | View Details | [359..522] | 28.0 | PhoP receiver domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)