













| General Information: |
|
| Name(s) found: |
int6 /
SPBC646.09c
[Sanger Pombe]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 501 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA eukaryotic 43S preinitiation complex [IDA cytosol [IDA eukaryotic translation initiation factor 3 complex [TAS] |
| Biological Process: |
ascospore formation
[IMP positive regulation of translational initiation [IC] eukaryotic translation initiation factor 3 complex assembly [IMP proteasomal ubiquitin-dependent protein catabolic process [IGI protein complex assembly [IMP chromosome segregation [IMP response to caffeine [IMP proteasome localization [IMP positive regulation of proteasomal ubiquitin-dependent protein catabolic process [IMP proteasome assembly [IMP cellular protein localization [IGI translation [IEA] |
| Molecular Function: |
translation initiation factor activity
[IEA]
protein binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSELKSTSP LAVKYDLSQK IMQHLDRHLI FPLLEFLSLR QTHDPKELLQ AKYDLLKDTN 60
61 MTDYVANLWT NLHGGHTDED MANAFAEKRR SVLQELSELE EEVQGILGVL ENPDLIAALR 120
121 QDKGQNLQHL QEHYNITPER IAVLYKFAQF QYNCGNYGGA SDLLYHFRAF SKDPELNASA 180
181 TWGKFASEIL TVDWDGAMEE LGKLREMVDS KSFKDSAVQL RNRTWLLHWS LFPLFNHANG 240
241 CDTLCDLFFY TPYLNTIQTS CPWLLRYLTV AVVTNQNNAN QKPRNPRQSY QRRMRDLVRI 300
301 ISQENYEYSD PVTSFISALY TEVDFEKAQH CLRECEEVLK TDFFLVSLCD HFLEGARKLL 360
361 AEAYCRIHSV ISVDVLANKL EMDSAQLIQL VENRNNPSVA AASNVAADQS TEDESIESTS 420
421 TNVVADDLIT EAETATEAEE PEPEVQFGFK AKLDGESIII EHPTYSAFQQ IIDRTKSLSF 480
481 ESQNLEQSLA KSISELKHAT V |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
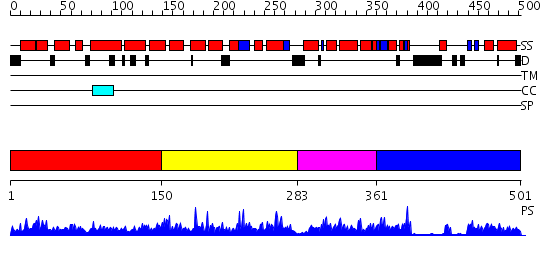
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..149] | 80.958607 | No description for PF09440.1 was found. No confident structure predictions are available. | |
| 2 | View Details | [150..282] | 2.033991 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [283..360] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [361..501] | 3.17 | Solution structure of the PCI domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)