













| General Information: |
|
| Name(s) found: |
rqh1 /
SPAC2G11.12
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1328 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear replication fork
[IC RecQ helicase-Topo III complex [IDA |
| Biological Process: |
reciprocal meiotic recombination
[IMP UV protection [IMP recombinational repair [IGI replication fork processing at rDNA locus [IGI double-strand break repair via homologous recombination [IGI chromosome segregation [IMP resolution of meiotic recombination intermediates [TAS telomere maintenance [IGI negative regulation of mitotic recombination [IMP response to DNA damage stimulus [IMP DNA unwinding involved in replication [ISS maintenance of rDNA [IMP resolution of mitotic recombination intermediates [IMP replication fork processing [TAS intra-S DNA damage checkpoint [IDA mitotic metaphase/anaphase transition [IMP |
| Molecular Function: |
ATP binding
[IC DNA binding [TAS protein binding [IPI ATP-dependent 3'-5' DNA helicase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTVTKTNLNR HLDWFFRESP QKIENVTSPI KTLDFVKVKV SSSDIVVKDS IPHKSKNVFD 60
61 DFDDGYAIDL TEEHQSSSLN NLKWKDVEGP NILKPIKKIA VPASESEEDF DDVDEEMLRA 120
121 AEMEVFQSCQ PLAVNTADTT VSHSTSSSNV PRSLNKIHDP SRFIKDNDVE NRIHVSSASK 180
181 VASISNTSKP NPIVSENPIS ATSVSIEIPI KPKELSNNLP FPRLNNNNTN NNNDNNAIEK 240
241 RDSASPTPSS VSSQISIDFS TWPHQNLLQY LDILRDEKSE ISDRIIEVME RYPFSSRFKE 300
301 WIPKRDILSQ KISSVLEVLS NNNNSNNNNG NNGTVPNAKT FFTPPSSITQ QVPFPSTIIP 360
361 ESTVKENSTR PYVNSHLVAN DKITATPFHS EAVVSPLQSN IRNSDIAEFD EFDIDDADFT 420
421 FNTTDPINDE SGASSDVVVI DDEEDDIENR PLNQALKASK AAVSNASLLQ SSSLDRPLLG 480
481 EMKDKNHKVL MPSLDDPMLS YPWSKEVLGC LKHKFHLKGF RKNQLEAING TLSGKDVFIL 540
541 MPTGGGKSLC YQLPAVIEGG ASRGVTLVIS PLLSLMQDQL DHLRKLNIPS LPLSGEQPAD 600
601 ERRQVISFLM AKNVLVKLLY VTPEGLASNG AITRVLKSLY ERKLLARIVI DEAHCVSHWG 660
661 HDFRPDYKQL GLLRDRYQGI PFMALTATAN EIVKKDIINT LRMENCLELK SSFNRPNLFY 720
721 EIKPKKDLYT ELYRFISNGH LHESGIIYCL SRTSCEQVAA KLRNDYGLKA WHYHAGLEKV 780
781 ERQRIQNEWQ SGSYKIIVAT IAFGMGVDKG DVRFVIHHSF PKSLEGYYQE TGRAGRDGKP 840
841 AHCIMFYSYK DHVTFQKLIM SGDGDAETKE RQRQMLRQVI QFCENKTDCR RKQVLAYFGE 900
901 NFDKVHCRKG CDICCEEATY IKQDMTEFSL QAIKLLKSIS GKATLLQLMD IFRGSKSAKI 960
961 VENGWDRLEG AGVGKLLNRG DSERLFHHLV SEGVFVEKVE ANRRGFVSAY VVPGRQTIIN1020
1021 SVLAGKRRII LDVKESSSKP DTSSRSLSRS KTLPALREYQ LKSTTASVDC SIGTREVDEI1080
1081 YDSQMPPVKP SLIHSRNKID LEELSGQKFM SEYEIDVMTR CLKDLKLLRS NLMAIDDSRV1140
1141 SSYFTDSVLL SMAKKLPRNV KELKEIHGVS NEKAVNLGPK FLQVIQKFID EKEQNLEGTE1200
1201 LDPSLQSLDT DYPIDTNALS LDHEQGFSDD SDSVYEPSSP IEEGDEEVDG QRKDILNFMN1260
1261 SQSLTQTGSV PKRKSTSYTR PSKSYRHKRG STSYSRKRKY STSQKDSRKT SKSANTSFIH1320
1321 PMVKQNYR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
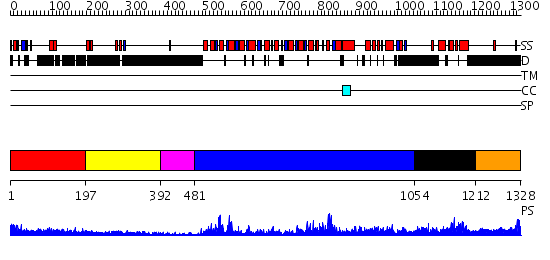
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..196] | 3.193997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [197..391] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [392..480] | 28.0 | No description for 2fdcA was found. | |
| 4 | View Details | [481..1053] | 94.39794 | No description for 2v1xA was found. | |
| 5 | View Details | [1054..1211] | 19.0 | E. coli RecQ HRDC domain | |
| 6 | View Details | [1212..1328] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)