













| General Information: |
|
| Name(s) found: |
fep1 /
SPAC23E2.01
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 564 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
cellular iron ion homeostasis
[TAS regulation of transcription from RNA polymerase II promoter in response to iron ion starvation [IMP gene-specific transcription from RNA polymerase II promoter [RCA regulation of iron ion transport [IMP |
| Molecular Function: |
DNA binding
[IDA transcription factor activity [IDA zinc ion binding [IEA] protein binding [IPI protein homodimerization activity [IDA specific RNA polymerase II transcription factor activity [RCA sequence-specific DNA binding [IC] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAKPAPFGQ SCSNCHKTTT SLWRRGPDNS LLCNACGLYQ KHRKHARPVK SEDLKDISPL 60
61 IQQVCKNGTC AGDGFCNGTG GSASCTGCPA LNNRIRSLNA SKSQSGRKSL SPNPSSVPSS 120
121 TETKASPTPL ESKPQIVSDT TTETSNGTSR RRSSHNQHED SSPPHEPSVT FCQNCATTNT 180
181 PLWRRDESGN PICNACGLYY KIHGVHRPVT MKKAIIKRRK RLVFNGNANE SQHNLKRMSS 240
241 GDSGSSVKQQ STRDGPFSKS FPNGNGHASG NSGEGLAEHG MNTGVLPPAS TFPSYNSNFT 300
301 GFLPSSFNPS PLMTLSRLAA GEPDNNGKVY YSYGPTQEQS ILPLPENKHE GLPPYQNEYV 360
361 PNGIRANQVV YPGQLVAVGN DSSKQLSEST TSNTDNNGVA TANQSNPLGM KFHLPPILPV 420
421 GESVCLPPRT SAKPRIAEGI ASLLNPEEPP SNSDKQPSMS NGPKSEVSPS QSQQAPLIQS 480
481 STSPVSLQFP PEVQGSNVDK RNYALNVLSQ LRSQHDLMIQ ELHNLNQHIQ QIDEWLRSSD 540
541 NENMASEHIK SSTPAVVASG ALQT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
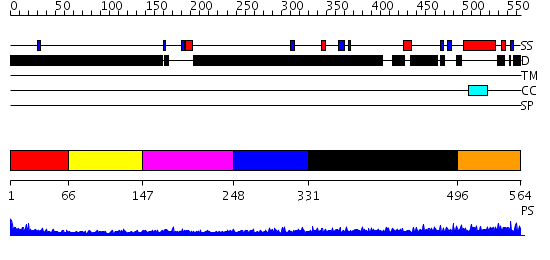
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..65] | 2.76 | Erythroid transcription factor GATA-1 | |
| 2 | View Details | [66..146] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [147..247] | 5.69897 | Erythroid transcription factor GATA-1 | |
| 4 | View Details | [248..330] | 1.185991 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [331..495] | 5.185985 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [496..564] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)