













| General Information: |
|
| Name(s) found: |
exo2 /
SPAC17A5.14
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1328 amino acids |
Gene Ontology: |
|
| Cellular Component: |
perinuclear region of cytoplasm
[IEA]
cytoplasm [IDA cytoplasmic mRNA processing body [ISS cytosol [IDA |
| Biological Process: |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay
[IEA]
rRNA processing [ISS mRNA catabolic process [ISS karyogamy [IEA] |
| Molecular Function: |
magnesium ion binding
[IDA [NOT]ribonuclease H activity [IDA RNA binding [IEA] double-stranded DNA specific 5'-3' exodeoxyribonuclease activity [IDA single-stranded DNA specific 5'-3' exodeoxyribonuclease activity [IDA ribonuclease activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGIPKFFRWM SERYPLCSQL IENDRIPEFD NLYLDMNGIL HNCTHKNDDH SSPPLPEEEM 60
61 YIAIFNYIEH LFEKIKPKKL LYMAVDGCAP RAKMNQQRSR RFRTAKDAHD ARLKAERNGE 120
121 DFPEEQFDSN CITPGTTFME RVSRQLYYFI HKKVTNDSQW QNIEVIFSGH DCPGEGEHKI 180
181 MEYIRTQKAQ PSYNPNTRHC LYGLDADLIM LGLLSHDPHF CLLREEVTFG PASRNRSKEL 240
241 AHQKFYLLHL SLLREYLEFE FQECRSTFTF KYDLEKILDD FILLAFFVGN DFLPHLPGLH 300
301 INEGALALMF SIYKKVMPSA GGYINEKGVI NMARLELILL ELENFEKEIF KAEVSETKNN 360
361 GNSDKPSFDF LKYITESTND IKAMTGEQKN YFLQIKKFLS SREPFIDFSA NISSVDQRFL 420
421 RRLCNDLHLS FSKIIKVDGT HLLRITFRDL EFNDEDEDEI EQDEIERVLQ KYDNIPLLNE 480
481 EQALKEKNVE KDFIQWKDDY YRSKVGFSYY DEEALKAMAE RYVEGLQWVL FYYYRGCQSW 540
541 GWYYNYHFAP KISDVLKGLD VKIDFKMGTP FRPFEQLMAV LPARSQALVP PCFRDLMVNS 600
601 ESPIIDFYPE NFALDQNGKT ASWEAVVIIP FIDETRLIDA LASKDKFLTE EERKRNSFNA 660
661 PTVFSLAEDY TSFYPSSLPS LFPDLVTRCI QKPYSLPSME GKEYLVGLCP GVFLGAFGMV 720
721 GFPSFHTLKH KAELVYHGIN VFGNESRNPS VIVNVEDVKS ALTSEQIAMQ YVGKRIFVDW 780
781 PYLREAYVES AMDESYMYLA SNSTIEKRDL AEIEKSQWGR KCSHKIREYS KRFGVLFGDI 840
841 SLLLQVRPIK GLEYTREGAL VKIFNESVLE DYPAQLVVEK IAIDDPRFTE REAPPVEVEY 900
901 PPGTKAFHLG EYNYGRPAQI TGCKDNKLII WLSTAPGLDA QWGRVLVNDS KSKEKYYPSY 960
961 IVAKLLNIHP LLLSKITSSF LISNGTKREN IGLNLKFDAR NQKVLGFSRK STKGWEFSNK1020
1021 TVALVKEYIN TFPQLFNILT THATKDNLTV KDCFPKDDTQ QLAAVKHWIK EKGINSLTRV1080
1081 SLDEDALDSD IIKLIEEKAS TIDSTYQVPK KVFGVPRYAL LKPSQTRGIL HSQEFALGDR1140
1141 VVYVQDSGKV PIAAYGTVVG IMLHHLDVVF DLPFMSGTTL DGRCSPYHGM QVEVSMVLNV1200
1201 TNPQFVVNTR AGKNRKTNVS ANNVSQGTDS RLVTKPTSTF PSPPSPPSSS VWNKREHHPK1260
1261 PFSLHQVPPP ESLIHKSKSK FSKGNHHSTN GTQSIRGRGG KRGKPLRSKE LNRKHDHIVQ1320
1321 PMGKLQIN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
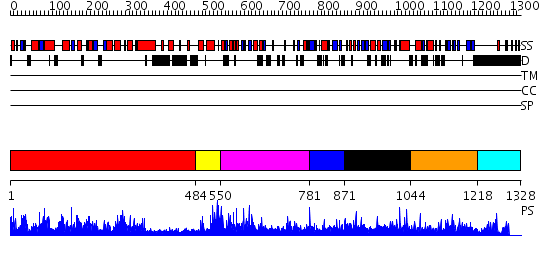
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..483] | 1.15 | Crystal structure of the human FEN1-PCNA complex | |
| 2 | View Details | [484..549] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [550..780] | 2.087951 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [781..870] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [871..1043] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1044..1217] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1218..1328] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)