













| General Information: |
|
| Name(s) found: |
cut9 /
SPAC6F12.15c
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 671 amino acids |
Gene Ontology: |
|
| Cellular Component: |
anaphase-promoting complex
[IDA cytosol [IDA |
| Biological Process: |
cyclin catabolic process
[IC DNA-dependent DNA replication [IMP anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process [IC mitotic metaphase/anaphase transition [IMP protein ubiquitination [IC |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVVKRTQTDS RMQSTPGNHN HPDAHANAAY MTPPSMGALN ANNSNSQLST LTISPMTYLA 60
61 NNTSTDGSFL KERNAQNTDS LSREDYLRLW RHDALMQQQY KCAAFVGEKV LDITGNPNDA 120
121 FWLAQVYCCT GDYARAKCLL TKEDLYNRSS ACRYLAAFCL VKLYDWQGAL NLLGETNPFR 180
181 KDEKNANKLL MQDGGIKLEA SMCYLRGQVY TNLSNFDRAK ECYKEALMVD AKCYEAFDQL 240
241 VSNHLLTADE EWDLVLKLNY STYSKEDAAF LRSLYMLKLN KTSHEDELRR AEDYLSSING 300
301 LEKSSDLLLC KADTLFVRSR FIDVLAITTK ILEIDPYNLD VYPLHLASLH ESGEKNKLYL 360
361 ISNDLVDRHP EKAVTWLAVG IYYLCVNKIS EARRYFSKSS TMDPQFGPAW IGFAHSFAIE 420
421 GEHDQAISAY TTAARLFQGT HLPYLFLGMQ HMQLGNILLA NEYLQSSYAL FQYDPLLLNE 480
481 LGVVAFNKSD MQTAINHFQN ALLLVKKTQS NEKPWAATWA NLGHAYRKLK MYDAAIDALN 540
541 QGLLLSTNDA NVHTAIALVY LHKKIPGLAI THLHESLAIS PNEIMASDLL KRALEENSLT 600
601 SGFLNSKYVF EDEVSEYMQQ SNLNTSDKSM SMEDQSGKVT ESVNDRTQVL YADSRSEMMM 660
661 DDIEGNVSEQ R |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Lid1-TAP APC complex on iontrap. | Unpublished Data |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
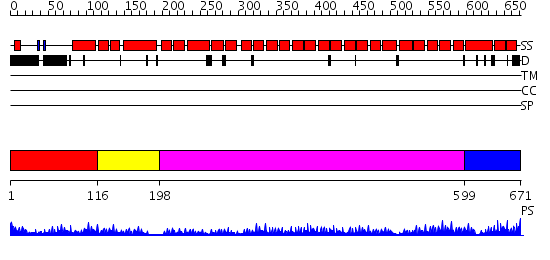
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..115] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [116..197] | 24.522879 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 3 | View Details | [198..598] | 59.0 | The superhelical TPR domain of O-linked GlcNAc transferase reveals structural similarities to importin alpha. | |
| 4 | View Details | [599..671] | 17.522879 | Designed protein cTPR3 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)