













| General Information: |
|
| Name(s) found: |
cut23 /
SPAC6F12.14
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 565 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA anaphase-promoting complex [TAS spindle pole body [IDA |
| Biological Process: |
regulation of mitotic metaphase/anaphase transition
[IEA]
anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process [IC mitotic metaphase/anaphase transition [IMP protein ubiquitination [IC |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTVSLNTMMG VDGLEDQEQL REIRNCLLKC ISECSERGLV YAVRWAAEML NGMNPIEMEH 60
61 IPFSSTPTGE FDLDPDMANE KLLEVEEKNI YLLAKSYFDC KEFERAAYTL QNCKSSKSIF 120
121 LRLYSKYLAG EKKSEEENET LLNTNLTLSS TNREFYYISE VLESLHYQGN KDPYLLYLSG 180
181 VVYRKRKQDS KAIDFLKSCV LKAPFFWSAW LELSLSIDSL ETLTTVVSQL PSTHIMTKIF 240
241 YVYASHELHQ VNSSAYEKLA EAEIIFPNSR YLKTQRALLT YDSRDFDEAE SLFENILTND 300
301 PYRLDDMDTY SNVLFVLENK SKLGFLAQVA SSIDKFRPET CSIIGNYYSL LSEHEKAVTY 360
361 FKRALQLNRN YLSAWTLMGH EYVELKNTHA AIESYRLAVD VNRKDYRAWY GLGQTYEVLD 420
421 MHFYALYYFQ RATALRPYDQ RMWQALGNCY EKIDRPQEAI KSYKRALLGS QTNSSILVRL 480
481 GNLYEELQDL NSAASMYKQC IKTEETEISP ETIKARIWLA RWELGKKNYR EAELYLSEVL 540
541 NGDLELEEAK ALLRELRSRM EHSYD |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Lid1-TAP APC complex on iontrap. | Unpublished Data |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
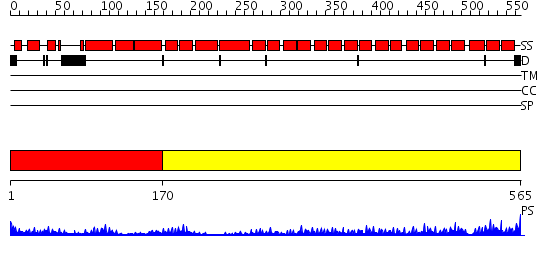
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..169] | 7.69897 | Crystal Structure of FKBP52 C-terminal Domain complex with the C-terminal peptide MEEVD of Hsp90 | |
| 2 | View Details | [170..565] | 48.221849 | The superhelical TPR domain of O-linked GlcNAc transferase reveals structural similarities to importin alpha. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)