













| General Information: |
|
| Name(s) found: |
cnd1 /
SPBC776.13
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1158 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA inner kinetochore of condensed chromosome [IDA cytoplasm [IDA nucleolus [IDA condensin complex [IDA |
| Biological Process: |
mitotic chromosome condensation
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLDLLSRLK KYIHDEENSD LDSIYAECEN AGLTAVVNDV IDTLLLGTSS CFDEDCLEKL 60
61 FAICSHFADL SSSVRNKVYD LLTSNISSES AILEDMISAN ATDFTVPQTN LETTGIAFQL 120
121 TVNSLSSSNQ LSVIRSSTNT VKGRKKNPTT NSNWNGISHV NALLDAIITL FQKKLSRVWT 180
181 TSSERDMFLS LFLKPIYTLM ESEINIKNAS FRSRLFNIIG LAVQFHNHTT AAETNIIQNL 240
241 QYFEHLSEYA ADLVHIVTVQ FNSVTLAEGI IRTLCSLEFN DNDVKGPKQV ALFLVRLSSL 300
301 IPNLCLKQLT QLVKLLDSES YTLRCAIIEV LANVVIDQIH DEAQNEMSES VPATVQSLMD 360
361 LLSERLLDIS PYCRTKVLHV FIKIFDLPIK YPRKRQEIAE LVIRCLQDRS SHVRRNAIKL 420
421 FSKLLTTHPF SVMHNGLLTR NIWEKGLSII EEQLNSLQPK QQEKVVDSEL EVDENLLEDA 480
481 TMIQDDESHE GESHLENSLS EYVDSVPAEE IVKVNLTKRF YLEALQYIDI VEAGAKIISQ 540
541 LLFAKNKSEV IESMDFFVFC NSFGISSSKL YIKKMIHLIW VKGTSDEGNN IQNHVLSCYK 600
601 TLFFEPPPNS GTNEAANYIA RNLISLTYDA SLAELTSLEQ MLCILMKDGY FSHLVITKLW 660
661 QVYSYQKKDI SRTQRRGSII VIGMLALGNT DVVMQGLDHL IQIGLGPPGL EDLVLARYTC 720
721 IAIKRIGKDA SGSSNINFPN SHTLCQKLCM LLLRPSFSEE WFGLEEQAIE AIYAVAKHPD 780
781 ELCTNIILLL TKQLFKPSNH ENTTSNDDHA MDEDLDDSPE EETLKDEEEI GIRLAHLIFL 840
841 VGHVAIKQLV YIEYCEAEFK RRKADAERLA VQNSNNPING QETSEYDLIT GTSEDDFSEA 900
901 MTFIRERELL YGENSLLSRF APLVVELCSN HKSHNNQSLL LAASLTLSKF MCLSNNFCME 960
961 HLPLLITILE KCDNPIIRNN LVIGLADLTV CFNHFIDEIS EYLYRRLMDE ESSVKKTCFM1020
1021 TLAFLILAGQ IKVKGQLGIM ARSLEDEDAR ISDLARMFFT DFAAKDNSVY NNFIDIFSVL1080
1081 SRSAEEQDED DAKFKRIIRF LTSFIEKERH TKQLAERLAA RLDRCKTQRQ WDHVVYALSL1140
1141 LPHKADNIQK LIDDGYHE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
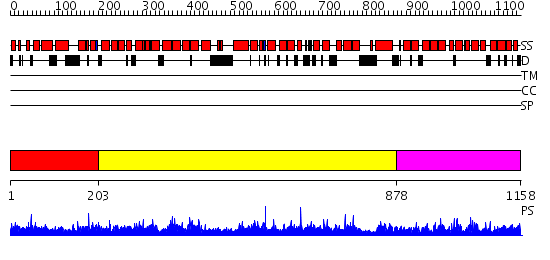
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..202] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [203..877] | 2.28 | Adaptin beta subunit N-terminal fragment | |
| 3 | View Details | [878..1158] | 60.221849 | Adaptin beta subunit N-terminal fragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)