













| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSPKQEEYEV ERIVDEKLDR NGAVKLYRIR WLNYSSRSDT WEPPENLSGC SAVLAEWKRR 60
61 KRRLKGSNSD SDSPHHASNP HPNSRQKHQH QTSKSVPRSQ RFSRELNVKK ENKKVFSSQT 120
121 TKRQSRKQST ALTTNDTSII LDDSLHTNSK KLGKTRNEVK EESQKRELVS NSIKEATSPK 180
181 TSSILTKPRN PSKLDSYTHL SFYEKRELFR KKLREIEGPE VTLVNEVDDE PCPSLDFQFI 240
241 SQYRLTQGVI PPDPNFQSGC NCSSLGGCDL NNPSRCECLD DLDEPTHFAY DAQGRVRADT 300
301 GAVIYECNSF CSCSMECPNR VVQRGRTLPL EIFKTKEKGW GVRSLRFAPA GTFITCYLGE 360
361 VITSAEAAKR DKNYDDDGIT YLFDLDMFDD ASEYTVDAQN YGDVSRFFNH SCSPNIAIYS 420
421 AVRNHGFRTI YDLAFFAIKD IQPLEELTFD YAGAKDFSPV QSQKSQQNRI SKLRRQCKCG 480
481 SANCRGWLFG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
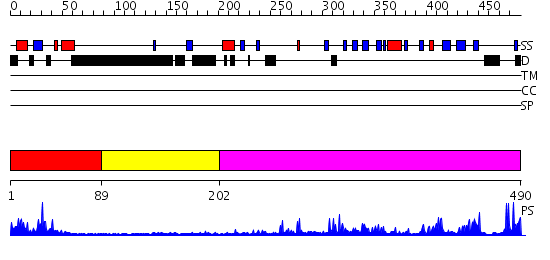
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..88] | 8.09691 | Histone methyltransferase clr4, chromo domain | |
| 2 | View Details | [89..201] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [202..490] | 62.522879 | SET domain of Clr4 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)