













| General Information: |
|
| Name(s) found: |
cid13 /
SPAC821.04c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 578 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA cytosol [IDA |
| Biological Process: |
DNA replication checkpoint
[IMP DNA integrity checkpoint [TAS mRNA polyadenylation [IDA |
| Molecular Function: |
ATP binding
[IEA]
RNA binding [IEA] polynucleotide adenylyltransferase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDNANCVGGC KFETRSFQYR RRIPYSLGAD PLPPVHPLSL KNLVDIDTDL ISSQLYELYD 60
61 SIILNDSGLE RRYAFVQKLE QILKKEFPYK NIKTSLFGST QSLLASNASD IDLCIITDPP 120
121 QCAPTTCEVS AAFARNGLKK VVCISTAKVP IVKVWDSELQ LSCDCNINKT ISTLNTRLMR 180
181 SYVLCDPRVR PLIVMIKYWA KRRCLNDAAE GGTLTSYTIS CMVINFLQKR DPPILPSLQM 240
241 LPHLQDSSTM TDGLDVSFFD DPDLVHGFGD KNEESLGILF VEFFRFFGYL FDYEHFVLSI 300
301 RHGTFLSKRA KGWQFQLNNF LCVEEPFHTS RNLANTADEI TMKGIQLEFR RVFRLLAYNC 360
361 NVDDACSQFT FPSLTDTSFM DDYVNELQLE IVPGFSHGRD SSDTSCTESP PEPSHFAWAF 420
421 DPYNATASPY YNQNINSSID YSSIYSNDVP AIPPNVPYTF VDPYTYACYI NNNSYLPPSY 480
481 MDFYTWYNSP YPKSSHHFDE RHGGDRHEKN LSNSRRYSRN KFHKKKQSSG PFQYYPDAFS 540
541 FTPTDNNSPP SNSSSSEVVS PVSLHSEPVL STVQAFKS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
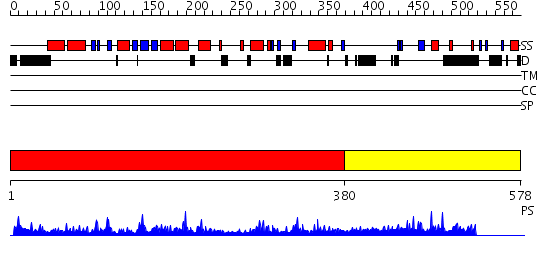
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..379] | 62.154902 | Poly(A) polymerase, middle domain; Poly(A) polymerase N-terminal, catalytic domain; Poly(A) polymerase, C-terminal domain | |
| 2 | View Details | [380..578] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)