













| General Information: |
|
| Name(s) found: |
cce1 /
SPAC25G10.02
[Sanger Pombe]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 258 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA |
| Biological Process: |
DNA recombination
[IMP mitochondrial genome maintenance [IMP |
| Molecular Function: |
magnesium ion binding
[IEA]
DNA binding [IMP endodeoxyribonuclease activity [IMP crossover junction endodeoxyribonuclease activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATVKLSFLQ HICKLTGLSR SGRKDELLRR IVDSPIYPTS RVLGIDLGIK NFSYCFASQN 60
61 EDSKVIIHNW SVENLTEKNG LDIQWTEDFQ PSSMADLSIQ LFNTLHEKFN PHVILMERQR 120
121 YRSGIATIPE WTLRVNMLES MLYALHYAEK RNSIEQKIQY PFLLSLSPKS TYSYWASVLN 180
181 TKASFSKKKS RVQMVKELID GQKILFENEE ALYKWNNGSR VEFKKDDMAD SALIASGWMR 240
241 WQAQLKHYRN FCKQFLKQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
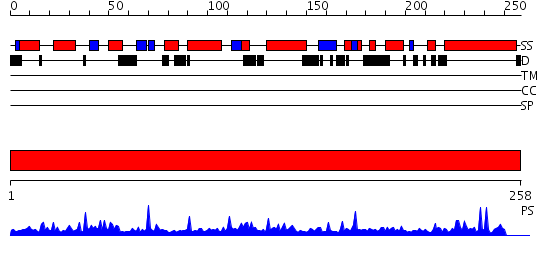
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..258] | 112.0 | Mitochondrial resolvase ydc2 N-terminal domain; Mitochondrial resolvase ydc2 catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)