













| General Information: |
|
| Name(s) found: |
Cap-PA /
FBpp0074065
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1200 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cohesin complex
[ISS][NAS]
|
| Biological Process: |
sister chromatid cohesion
[ISS]
|
| Molecular Function: |
ATP binding
[IEA]
protein binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHIKQIIIQG FKSYKDQTVV EPFDKRHNVV VGRNGSGKSN FFYAIQFVLS DEFTHLRPEQ 60
61 RQSLLHEGTG ARVISAYVEI IFDNSDNRVP IDKEEIFLRR VIGAKKDQYF LNKKVVPRNE 120
121 VVNLLESAGF SSSNPYYIVK QGKINQMATA ADSYRLKLLR EVAGTRVYDE RKEESLNLLR 180
181 ETDSKVEKIS EYLKTIEDRL QTLEEEKEEL KEYQKWDKTR RTLEYIRYET ELKDTKKALD 240
241 ELQLQRKSSS DKKKIYNIEI QKAQEKIKDV QKNLKEAKKK VQSTKEERSV LMTEQQQLLR 300
301 EKTKLDLTIV DLNDEVQGDN KSKERADQEL KNLKVTIAER EKELDDVKPK YEAMKRKEED 360
361 CSRELQLKEQ KRKELYAKQG RGSQFSSRED RDKWITNELK SISKQTRDKI AHHAKLVEDL 420
421 KKDATSEKDL GQKIEEHSSE LEQLRLQIDE HNKKYYELKK TKDQHQSMRN ELWRKETQMT 480
481 QQLQTHKEEL SRADQALRSM AGKPILNGCD SVRKVLDSFV ERGGQSAEIA RAYYGPVIEN 540
541 FSCDKTIYTA VEVTAANRLF HHIVESEYEG TQILKEMNKL KLPGEVTFMP LNRLQVKIHD 600
601 YPDDPDSIPM ISKLKYDEQH DKALRYIFGK TLICRNLERA TELAKSTGLD CVTLDGDQVS 660
661 SKGSLTGGYF NTSRSRLEMQ KKRTEYTSQI AEFEKKLSKL RNELKSTENN INSIVSEMQK 720
721 TETKQGKSKD VFEKVQGEIR LMKEELVRIE QYRAPKERSL AQCKASLESM TSTKSSLEAE 780
781 LKQELMSTLS SQDQREIDQL NDDIRRLNQE NKEAFTQRMQ FEVRKNKLDN LLINNLFRRR 840
841 DELIQALQEI SVEDRKRKLN NCKTELVSAE KRIKKVNSDL EEIEKRVMEA VQLQKELQQE 900
901 LETHVRKEKE AEENLNKDSK QLEKWSTKEN MLNEKIDECT EKIASLGAVP LVDPSYTRMS 960
961 LKNIFKELEK ANQHLKKYNH VNKKALDQFL SFSEQKEKLY RRKEELDIGD QKIHMLIQSL1020
1021 EMQKVEAIQF TFRQVAQNFT KVFKKLVPMG AGFLILKTKD NEGDEMEKEV ENSDAFTGIG1080
1081 IRVSFTGVEA EMREMNQLSG GQKSLVALAL IFSIQKCDPA PFYLFDEIDQ ALDAMHRKAV1140
1141 ANMIHELSDT AQFITTTFRP ELLENAHKFY GVRFRNKVSH IDCVTREEAK DFVEDDSTHA1200
1201 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
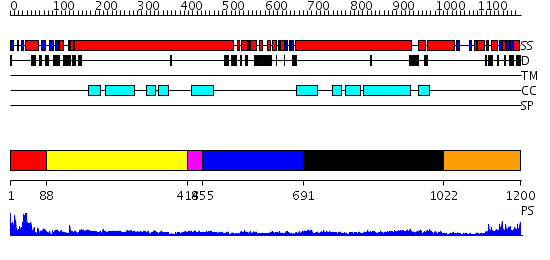
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | 39.522879 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. | |
| 2 | View Details | [88..417] | 11.154902 | Heavy meromyosin subfragment | |
| 3 | View Details | [418..454] | 4.39794 | No description for 2i1jA was found. | |
| 4 | View Details | [455..690] | 4.69 | Smc hinge domain | |
| 5 | View Details | [691..1021] | 2.42 | Tropomyosin | |
| 6 | View Details | [1022..1200] | 16.69897 | Structural biochemistry of ATP-driven dimerization and DNA stimulated activation of SMC ATPases. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)