













| General Information: |
|
| Name(s) found: |
mei-41-PA /
FBpp0074047
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2517 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
regulation of cell shape
[IMP]
DNA repair [TAS] mitotic cell cycle checkpoint [TAS] reciprocal meiotic recombination [IMP] regulation of mitosis [IGI][IMP] chromosome condensation [IGI] response to radiation [TAS] imaginal disc development [TAS] actin filament organization [IMP] telomere maintenance [IGI] meiotic anaphase I [IMP] double-strand break repair via synthesis-dependent strand annealing [IMP] regulation of syncytial blastoderm mitotic cell cycle [TAS] response to DNA damage stimulus [IMP] cellularization [TAS] meiosis [TAS] DNA damage checkpoint [IMP][TAS] cell cycle checkpoint [TAS] embryonic development via the syncytial blastoderm [TAS] oogenesis [TAS] double-strand break repair via single-strand annealing [IMP] female meiosis chromosome segregation [IMP] phosphorylation [NAS] |
| Molecular Function: |
protein kinase activity
[NAS]
protein serine/threonine kinase activity [IEA] binding [IEA] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTQRKDMWK LLYNHVNRNV SNFSGVYSVI EDILCQEPSL ISCSLVRELH NKFQDTFLLW 60
61 LLNKLAKCLS ESPDSSECIN LQRKILSSCC SNHPKLFERL VLAYVEAIEE THLQLSSLDL 120
121 GQLSNERKPA ITVRIFRCDV ECLQEFDPHC AIEDIKVPLE QADMYAKSLL EVLQHAHHIG 180
181 YATHGDIFSG SLHQALLILK ECDMDTKLAS LNYCHNVLRS QSASSWITNP DVGHYAQLTL 240
241 EATAIMWSAV AKWLDMGCMT RQELKRLNIT TKLLLEVLHM RARPAHHLGY LLLNEILSLP 300
301 TAIELDDGLL ETLSSYIQGQ LEHSVVPLEQ LVHLQQLMLS HWHCHPTHLV PILALMGLKQ 360
361 TEMRSGVVQV LTQSLVEILK KEEVLSKDWQ KLIAILRGFK QLEKLILSQS QHKIAEHEGH 420
421 IDSSVLAMLP LQCEIIKVAD TNWNNLSMQL VELESKCSAD RRHIHLEICS LLMQITFIRH 480
481 FLKTQTQHQL LAILQRHLKL SYLCAIRLET PSSVHTQMQS FYAQQYMRLF QSEETQEIFC 540
541 SNLPQLYISG FIKPEQLMKA LPTINNRSGR AQVIRLLLCS QPGKLSVFKV KDRIELYCPK 600
601 CRPLPKKLPG IYLGKCKQQL PCPDFSSTNL EMIANDLLFY PDFECIAQHL DLLCFEPNVI 660
661 LGLLRETEAL QKVSVKVIGQ LVSAMRVRSP EFLEQLANLV LAAIKAMLAK PLTEQNVLQQ 720
721 RSMLNVLTAI AHMEDNEIWL FHWFKMTFFF LVHTRSLVAQ EAVLAATEMC ASQGLQTIHL 780
781 WNWYKRDALD LTVRLALNVY LLDGVRFTRS LRALTKMLGF TCVQEFTCKY HRLLTAMVLP 840
841 HCIVNPLCKG VLVLIAKQMQ KHIGTLFSIS FLRIYTHVFL TEEPELANSC IELVVSCTQS 900
901 SLQQLMNADV KQTVAELLIY FNRNPTFVMR SFQSLLQLSI GSLEELSSQT ANAEFANFIA 960
961 ERFLGVITYF ESCLSEPSFE KPLKEETLYS LGQIMRFVGS QHVTQFRFKI IAMLSFVHTL1020
1021 QEPRLQRICL KIWHIFLHVV NVQELGPSLG RIVATLQPLL ADNESVKQVN DLYEFIILRN1080
1081 ASMLGTFITD LYFLDRMENV SPSIQKCIRR HTAHLDLKGL AEEENQSPPL VDQLRFLQKH1140
1141 ITDECLQVRV YALQHLGDLF GRRRPKLNST ILSELPLEPM LEQIVNVLMA GCQHDDSQLQ1200
1201 MASAKCLGEL GAIDASYLPS NYNFASPQHL PLNILSDDFA VLALTSLCRG YQFQQNTKHV1260
1261 DSFSLAIQET LAICGISPKE QKKVQLWQSL PARMRQLMEP MLHSCYTCVH RPSTCLQQPL1320
1321 FGSHYSHNYY EEWAFLWASR LIDYLPSSGR RHLLSSYKPC IKRDSNMLST FYPYILLHAL1380
1381 LECTTEQRNH IQEEFMAVLQ ANEESSSSVR GRQELGAIKE NAFKQFESRK YAAGIKPLAS1440
1441 TLVSDRKEDS SRVPRLAGKL CAELLDFLQR WLREWQRIHG RSTGGKPPET IDSNYRKIHE1500
1501 FLNLIPKLLV SRASYNCGEY ARALSYLESY LEEGEDKSQR LLEQFTFLVE VYGSLRDPDS1560
1561 VEGAVQVRSY DMSVTQDILV NRLVERQQDM ITSYEQLLSS TDQMQPDHVR AMIDAYLRDT1620
1621 PKTAQLIADG LWQRLSDRYS DQCFAECKSE LLWRLGSYDE MEELQSNWPA QCSQGCLKLR1680
1681 RPLTTRIEFD SLLDGMRESV LEELRSCSAV QQHSYANAYD AVLKLHLVHE LHCSQELVEK1740
1741 LEQDRDEDNQ EKLMKNYFDD WQYRLQIVQP QVRIQESIYS FRRNILAELQ RRLTDRNHLL1800
1801 PHLKTELARI WLNSAQINRN AGQLQRAQLY ILKAAEYQPS GLFIERAKLL WQKGDQVMAM1860
1861 NYLEEQLSIM RSGCQGNVKQ LAAEQRHLFF RGKYLQAVYS AESMHLCADA VLKYFQEAIA1920
1921 VHRQSESCHV QMAQFLEKIL EARQGGKSEP TGEQDDMLIN VMVNYAKSLR YGSEHVYQSM1980
1981 PRLISLWLDT TESSTNTEQV KKMNDLLTNC CTALPTAVFY TVYSQMLSRL CHPVNDVFTV2040
2041 LRNVIIKLVE AYPQQSLWML LPHFKSAKAH RIKRCKLVLT DSRLQNSTFQ KLLQDFNSLT2100
2101 ERLMDLTNKE VTLDRTYKLS DLDTRLSKLC KQPEFSQILL PFEKYMQPTL PLNSDSNSSE2160
2161 GSHLPANQST VNWFPYQQIY ISGFQESVLI LRSAAKPKKL TIRCSDGKDY DVLVKPKDDL2220
2221 RRDARLMEFN GLVKRYLHQD APARQRRLHI RTYAVLPFNE ECGLVEWLPN LASYRSICMN2280
2281 LYAQRRLVMS TRQLQSLAVP LHESIERKRE VFTKQLVPAH PPVFQEWLRQ RFATPHSWYE2340
2341 ARNTYIRTVA VMSMVGYILG LGDRHGENIL FAEGNGDAVH VDFNCLFNQG ELLPYPEVVP2400
2401 FRLTHNMIVA MGPLGVEGSF RKCCEITLRL LKQESKTLMS ILRPFVYDVG AQTRKGAATA2460
2461 KITKDVQRIA DRLQGHVKRQ QANSIPLSTE GQVNFLINEA TKVDNLASMY IGWGAFL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
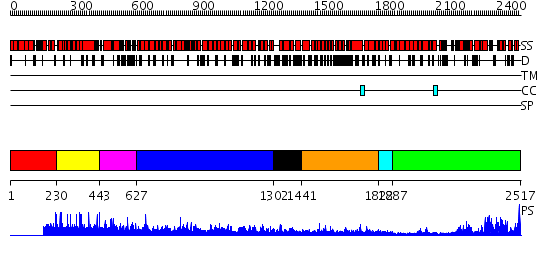
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..229] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [230..442] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [443..626] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [627..1301] | 9.0 | Crystal Structure of The Cand1-Cul1-Roc1 Complex | |
| 5 | View Details | [1302..1440] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1441..1821] | 15.09691 | Heavy meromyosin subfragment | |
| 7 | View Details | [1822..1886] | 6.221849 | Tropomyosin | |
| 8 | View Details | [1887..2517] | 69.39794 | Phoshoinositide 3-kinase (PI3K) helical domain; Phoshoinositide 3-kinase (PI3K); Phoshoinositide 3-kinase (PI3K), catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)