













| General Information: |
|
| Name(s) found: |
rdgB-PD /
FBpp0099561
[FlyBase]
rdgB-PC / FBpp0099560 [FlyBase] rdgB-PA / FBpp0073653 [FlyBase] |
| Description(s) found:
Found 46 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1259 amino acids |
Gene Ontology: |
|
| Cellular Component: |
subrhabdomeral cisterna
[NAS][TAS]
integral to plasma membrane [ISS] integral to membrane [ISS][TAS] plasma membrane [ISS] intracellular [IEA] |
| Biological Process: |
photoreceptor cell maintenance
[NAS]
sensory perception of smell [IMP] phosphoinositide metabolic process [IMP] rhodopsin mediated signaling pathway [IMP] visual perception [NAS] phototransduction [IMP][TAS] deactivation of rhodopsin mediated signaling [IMP][TAS] transport [IEA] |
| Molecular Function: |
metal ion binding
[IEA]
calcium ion binding [ISS] phosphoinositide binding [TAS] phosphatidylinositol transporter activity [IDA][NAS][TAS] calcium-transporting ATPase activity [ISS] phosphatidylcholine transmembrane transporter activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLIKEYRIPL PLTVEEYRIA QLYMIAKKSR EESHGEGSGV EIIINEPYKD GPGGNGQYTK 60
61 KIYHVGNHLP GWIKSLLPKS ALTVEEEAWN AYPYTRTRYT CPFVEKFSLD IETYYYPDNG 120
121 YQDNVFQLSG SDLRNRIVDV IDIVKDQLWG GDYVKEEDPK HFVSDKTGRG PLAEDWLEEY 180
181 WREVKGKKQP TPRNMSLMTA YKICRVEFRY WGMQTKLEKF IHDVALRKMM LRAHRQAWAW 240
241 QDEWFGLTIE DIRELERQTQ LALAKKMGGG EECSDDSVSE PYVSTAATAA STTGSERKKS 300
301 APAVPPIVTQ QPPSAEASSD EEGEEEEDDD EDENDAIGTG VDLSANQGGS AQRSRSQSIQ 360
361 MAQKGKFGSK GALHSPVGSA HSFDLQVANW RMERLEVDSK SNSDEEFFDC LDTNETNSLA 420
421 KWSSLELLGE GDDSPPPHGG PSSAASVGGR GNSRQEDSIF NQDFLMRVAS ERGNKRQLRS 480
481 SASVDRSHDS SPPGSPSTPS CPTTILILVV HAGSVLDAAS ELTAKKSDVT TFRGSFEAVM 540
541 RQHYPSLLTH VTIKMVPCPS ICTDALGILS SLSPYSFDAS PSAADIPNIA DVPIGAIPLL 600
601 SVASPEFHET VNKTVAAANI VYHEFLKSEE GHGFSGQIVM LGDSMGSLLA YEALCRSNGS 660
661 QPGTASGASN SGGDAATNIN THNPLSPRNS RLDDDERFIE ADLDAKRLLV APSPRRRRSS 720
721 SSSDSRATKL DFEVCDFFMF GSPLSVVLAA RKLHDAKAAL PRPNCHQVYN LFHPTDPIAS 780
781 RLEPLLSARF SILAPVNVPR YAKYPLGNGQ PLHLLEVIQS HPQHFNDGNN LLAGRRLSDA 840
841 SMQSTISGLI ENVSLSTIHA LQNKWWGTKR LDYALYCPEG LSNFPAHALP HLFHASYWES 900
901 PDVIAFILRQ IGKFEGIPFV GSNDDKDNAS FHPGQPREKW IKKRTSVKLK NVAANHRAND 960
961 VIVQEGREQR LNARFMYGPL DMITLHGEKV DVHIMKDPPA GEWTFLSTEV TDKNGRISYS1020
1021 IPDQVSLGYG IYPVKMVVRG DHTSVDCYMA VVPPLTECVV FSIDGSFTAS MSVTGRDPKV1080
1081 RAGAVDVCRH WQELGYLLIY ITGRPDMQQQ RVVSWLSQHN FPHGLISFAD GLSTDPLGHK1140
1141 TAYLNNLVQN HGISITAAYG SSKDISVYTN VGMRTDQIFI VGKVGKKLQS NATVLSDGYA1200
1201 AHLAGLQAVG GSRPAKGNAR MVIPRGCFNL PGQTANPRRR RYLERKTVSS CCLMVFQTT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
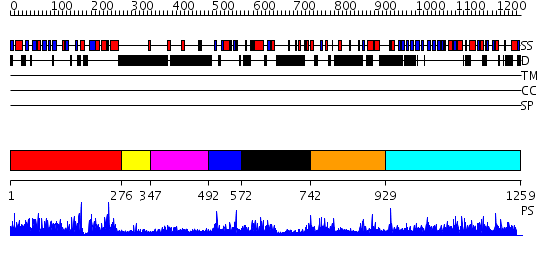
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..275] | 121.0 | Phoshatidylinositol transfer protein, PITP | |
| 2 | View Details | [276..346] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [347..491] | 2.09799 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [492..571] | 1.108986 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [572..741] | 1.111978 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [742..928] | 92.49485 | No description for PF02862.8 was found. No confident structure predictions are available. | |
| 7 | View Details | [929..1259] | 2.53 | No description for 2i33A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)