













| General Information: |
|
| Name(s) found: |
rap-PB /
FBpp0070655
[FlyBase]
rap-PA / FBpp0070654 [FlyBase] |
| Description(s) found:
Found 37 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 478 amino acids |
Gene Ontology: |
|
| Cellular Component: |
centrosome
[IDA]
centriole [IDA] spindle [IDA] |
| Biological Process: |
positive regulation of exit from mitosis
[IMP]
compound eye morphogenesis [IMP] cyclin catabolic process [IDA] protein catabolic process [IMP] eye-antennal disc morphogenesis [IMP] glial cell migration [IMP] |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFSPEYEKRI LKHYSPVARN LFNNFESSTT PTSLDRFIPC RAYNNWQTNF ASINKSNDNS 60
61 PQTSKKQRDC GETARDSLAY SCLLKNELLG SAIDDVKTAG EERNENAYTP AAKRSLFKYQ 120
121 SPTKQDYNGE CPYSLSPVSA KSQKLLRSPR KATRKISRIP FKVLDAPELQ DDFYLNLVDW 180
181 SSQNVLAVGL GSCVYLWSAC TSQVTRLCDL SPDANTVTSV SWNERGNTVA VGTHHGYVTV 240
241 WDVAANKQIN KLNGHSARVG ALAWNSDILS SGSRDRWIIQ RDTRTPQLQS ERRLAGHRQE 300
301 VCGLKWSPDN QYLASGGNDN RLYVWNQHSV NPVQSYTEHM AAVKAIAWSP HHHGLLASGG 360
361 GTADRCIRFW NTLTGQPMQC VDTGSQVCNL AWSKHSSELV STHGYSQNQI LVWKYPSLTQ 420
421 VAKLTGHSYR VLYLALSPDG EAIVTGAGDE TLRFWNVFSK ARSQKENKSV LNLFANIR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
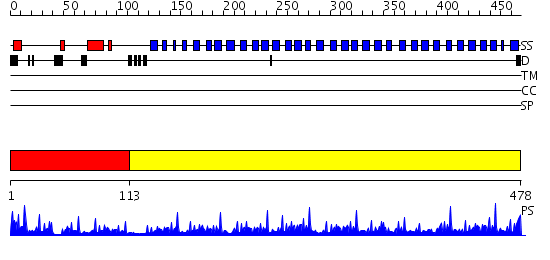
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..112] | 5.522879 | No description for 2ojhA was found. | |
| 2 | View Details | [113..478] | 77.154902 | PAF-AH Holoenzyme: Lis1/Alfa2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)