













| General Information: |
|
| Name(s) found: |
pon-PA /
FBpp0070649
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 670 amino acids |
Gene Ontology: |
|
| Cellular Component: |
basal part of cell
[TAS]
cytoplasm [IDA] cell cortex [IDA] basal cortex [NAS] |
| Biological Process: |
asymmetric neuroblast division
[TAS]
asymmetric protein localization [TAS] |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLETKSIAAL AFTETPRKRK RETLYKNAAP PPNPQQQPPE KDAQDAAVAS TESCFTNAAF 60
61 SSTPKKLMVA RRRLAKENSQ PNPFPGLEVK SIADLAQKQQ HQQHHQQQQQ QQQKLPTNPF 120
121 EVLRQPPKKK KREHACFENP GLNLELPEKQ FNPYEVVRSA TTPAKGFVNP ALNLRGSDAP 180
181 ASLNPFEIHR PSEASEAACN PSGVANPALA DRDSEEQLPT SLKIGLPFTP TLGCRIDFHG 240
241 MSLTQLTPSK LLAEKLVFSP VPAPKRSLGA ISEESSMDIG KELDRYQLEL ENSINEAKLR 300
301 KNGVLVDREL PRNSLEVELP KNTKVSLVME TNTQELMMQE VVTVDTQVER RLVCTRRRTL 360
361 TEISEASEVE EDTEKLLEQD REAEVILEQE KILEQEMVSE RERHLTREKQ LKQEKLLERE 420
421 KHLEREKLQE KLHEQLREKL QERAKHLEKE KLEEEELLER QLEEEREKEP TDGDVAYASE 480
481 SESDDEPDDL DFKAPARFVR AYRPAALPSK AASKESLQSI GSSKSAKSAE VKPSIGMKGM 540
541 IRKSIRRLMH PTSHTTPSEV KSEDKDEHGH GQGHGQHNIL NSIRHSLRRR PQKAAELEEQ 600
601 MEPVLADVSI IDTSERTMKL RSSVAQTEYM TIEQLTNEKK HSLRNSIRRS TRDVLRHVFH 660
661 KSHDAYATAK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
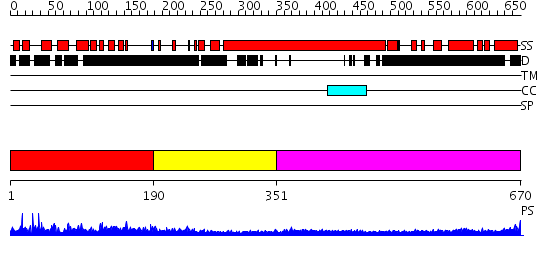
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..189] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [190..350] | 5.09691 | No description for 2h94A was found. | |
| 3 | View Details | [351..670] | 17.30103 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)