













| General Information: |
|
| Name(s) found: |
rod-PA /
FBpp0085156
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2089 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
kinetochore microtubule [IDA][NAS] spindle [IDA] kinetochore [IDA] outer kinetochore of condensed chromosome [NAS] |
| Biological Process: |
mitotic cell cycle spindle assembly checkpoint
[NAS][TAS]
mitotic sister chromatid segregation [IMP] |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MWSNFEASSL EDETISNIFS SRGIFQISSE EEVNKYPDVM ASVQCGKMVI GIDKSLLLLE 60
61 DELVAKCSFL AFGAAIEAIA ISSSGNLIVC GLSDGEVHGV FIKGLLLFSV AVNLEDVSLT 120
121 GGTFRSIHQL DQRFYFTCKN GSIYSLSDID ETCLETLGNM SLNENETIAC EKALLEDVTI 180
181 QRLSKGRSSC SVDCAVLLKR LLTGNISAEN NQDDLEAQPG LVITGDQSTM NFRNAANDVT 240
241 RINLPAQYGS LKQIFNLEHY IIALTDSGHL VEICPYTRTV YMCQTTQQKN LLIEDLLVME 300
301 CSEDNIELLV LTKSTAEGRF IKIVEYPTLN VKNEVEVPDH AWLVRQPKCA VNLYYLAAKE 360
361 VNQSSIPSVV EMMLVSETDP SDRFKKLICK GRLEEAEEFG KQFELCLQPI YEAKAKRILV 420
421 ELCNSNSQNV DTKFQDLLQL LSQVESKAFI KSHRMINLNS RHILERYLHE VKKRLTFEDD 480
481 EEDMLEIDEQ LHRLKTLAII DPYECNTDWQ KFIYDNNLVR MVKSLFNTDM PTACLIWRRH 540
541 SSSILPLLNE DELRTLLGFI PSNTKPFNVV QWLRQFIPMV SNTHPSIMPF ITDWSIEMTR 600
601 NLQYSPHWPD IGLEFCTKIL DIFEEIQFTY SDVRRQQERN VGKLRDLVNA LQDLSVLKTN 660
661 YNMGFTLDNY MQDSIDATAL SILQHVQLDK LQRLVKNFMY PIFQEKGRQP LDVIKQYISQ 720
721 LVASRLSSSS WLDRAMACIE LLHNEDSRLE CALSVLQNAP VPWPDTLAPL IRLRASTHPL 780
781 AVKINAEYEI QVIKIMKVKY GWPADSSDIN LELFMMRIVK LNLPDMLDDI SALTKAAPEL 840
841 STSANFNCCY QMARRGQVEM AYEFFKKLNG EKNSKSAKEV VEIMANILEN SSSASFESDI 900
901 KVQEHLNVLE LFKLFLPHVE SVYERRYLVI KHRFRLRKFG ILLSCSSELI PLHRRHCLLD 960
961 EAIERIIERA QATLNVSAFI ASEISELCTA LGLPKVFGLH RICQRIGCLP LSCALAYHVV1020
1021 SFVDCVPANA GEFVNLALEL LVQQIESAKG KNQETASSLQ LINENDPLSF YLAYELLTSA1080
1081 LLHESSRRHD LVELIKYIRV AVIHYPLDAI KSHYDGKEQA VNEHICRALD GISVALGETT1140
1141 INFSCALNGS FDVKTLTVPT TTKKRYSVSM FDEVEVQPIQ QQPQKKSVGG RMAIVKFLAR1200
1201 TLLLMVIETE PNNSLLMQVR NELPENTKAD VESACADFFL SLEHLTKVKE HDAWYVMSQY1260
1261 LLDFQKQNCR TKRIINEGFI TLQLRRIFRN AMGSKDVNFI ELFTMLVVDS EALALLEKLS1320
1321 VEVKTDLQKI NFLTLSAMYH EHIEDLDSVH MIRTKRLKLF YYLEFCQQDH RIKGKFNADM1380
1381 DNIEDLLKEF HNKQLDVQLL ERMSKDFGFD YQKILITQIL SILSAQELRF EVKRDTFGDE1440
1441 ELVMLSSAQE MRDMCQPYIN EINNVELFTS KLKHFIEDIN IYFYELYMCV INILMYFDSA1500
1501 PKEMEIWMNI LHFLRHKMIT RRRNRPGQVE TDTWLKSQRE NGVMPKISRY RLPFKPIVEQ1560
1561 PLKDILDSEL NVDNCQSWFP LIQMYTALKG SKDSSQNCDY FCMSAVKNSI SEYKSKNDSE1620
1621 SWSLHPTNNA FLQSILRLVE KVNNPNKAFL ILYFVSNYAR DGADQVEASY ECWKFVKEKG1680
1681 HLITDPKSRE QVAKVKRRYP IQKTQHLLYV YGLTDEKLLR QVENPTELIQ ALYHHELILK1740
1741 SSKVDINALV AEIAKLHDLC LATIQYRLLQ KWLALTMESA GDGTILEETF MEEQNWPEQA1800
1801 NGAESSNSQD ASENVNRAFY ILSSWPKAEA VKFLVGLIFK GGSVNTSTQL QMYECYSKLN1860
1861 DGSNSFTNTI SQRQFISIKC VHELKALGYK SNLEKFSDDH CNKIDILKMI WQRNAQNPLS1920
1921 LEVMANICLG FDIHLPQIWN GILKRMVMFH MVRELNALLD VLSCRPHLLH LDGLAQAWDY1980
1981 VLCHPLQNAV ELRSFKQEEV LHKTLLRLQG CPVVHALNLL QFAQLCIVVH RPHMAASLLS2040
2041 FCQSSEQRQK IKKMIHSHPV ENLRQQILDL EDAGILPVVL NFALKELNL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
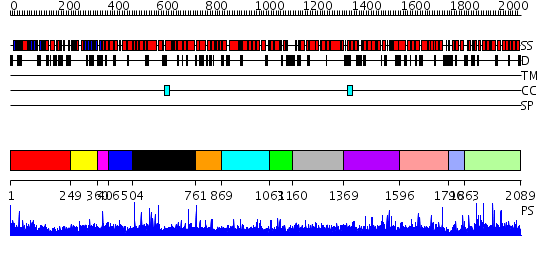
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..248] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [249..359] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [360..405] | 1.009989 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [406..503] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [504..760] | 1.010959 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [761..868] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [869..1062] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1063..1159] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1160..1368] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1369..1595] | 1.010963 | View MSA. No confident structure predictions are available. | |
| 11 | View Details | [1596..1795] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1796..1862] | N/A | No confident structure predictions are available. | |
| 13 | View Details | [1863..2089] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)