













| General Information: |
|
| Name(s) found: |
Sry-delta-PA /
FBpp0084924
[FlyBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 433 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][NAS]
polytene chromosome interband [IDA] |
| Biological Process: |
anterior/posterior axis specification, embryo
[IGI][IMP]
oogenesis [IGI][NAS] regulation of transcription, DNA-dependent [NAS] |
| Molecular Function: |
DNA binding
[IDA]
zinc ion binding [IEA] transcription activator activity [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTCFFCGAV DLSDTGSSSS MRYETLSAKV PSSQKTVSLV LTHLANCIQT QLDLKPGARL 60
61 CPRCFQELSD YDTIMVNLMT TQKRLTTQLK GALKSEFEVP ESGEDILVEE VEIPQSDVET 120
121 DADAEADALF VELVKDQEES DTEIKREFVD EEEEEDDDDD DEFICEDVDV GDSEALYGKS 180
181 SDGEDRPTKK RVKQECTTCG KVYNSWYQLQ KHISEEHSKQ PNHICPICGV IRRDEEYLEL 240
241 HMNLHEGKTE KQCRYCPKSF SRPVNTLRHM RMHWDKKKYQ CEKCGLRFSQ DNLLYNHRLR 300
301 HEAEENPIIC SICNVSFKSR KTFNHHTLIH KENRPRHYCS VCPKSFTERY TLKMHMKTHE 360
361 GDVVYGVREE APADEQQVVE ELHVDVDESE AAVTVIMSDN DENSGFCLIC NTNFENKKEL 420
421 EHHLQFDHDV VLK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
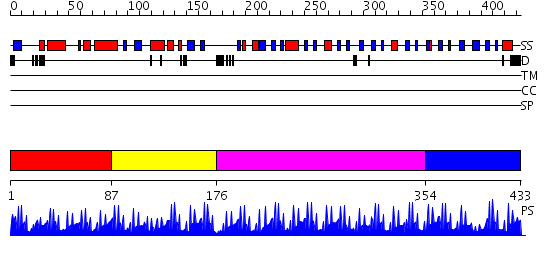
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..86] | 0.992 | Crystal structure of the zinc finger associated domain of the Drosophila transcription factor Grauzone | |
| 2 | View Details | [87..175] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [176..353] | 59.522879 | No description for 2i13A was found. | |
| 4 | View Details | [354..433] | 1.06 | Solution Structure of the N-terminal Zinc Fingers of the Xenopus laevis double stranded RNA binding protein ZFa |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)