













| General Information: |
|
| Name(s) found: |
CG7609-PB /
FBpp0084909
[FlyBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 776 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
peroxisome organization
[ISS]
|
| Molecular Function: |
peroxisome matrix targeting signal-2 binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPDSVEDTST ISLRICLEGH ANALALNKDN NQIALAGRSL LKVYSINSNG FTESCNMRGK 60
61 NQNLSYSAND VAWSTLDSNL LATAATNGVV SVWDLSKFGR QKQLLVYNEH ERTAHTVTFH 120
121 SSEPNILISG SQDGTIKCFD IRSDKSINTY FCNSESVRDV KFSPHTQNIF SAVSENGTVQ 180
181 LWDMRKWDKC MVQFTAHYGP VYTCDWHPTR NWLATGSRDK QIKVWNMDGR PGLEHTIHTI 240
241 AVVGRVKWRP ERTYHIASCA LVVDYSVHVW DIRRPYIPFA SFNEHTNVTT GIAWQGSDSH 300
301 CLLSISKDST IYKHAFKDAT RPALKANAQG ASLGRFGDIS FANKIKEYEP KTSGASNTKS 360
361 SSFIRRKTNV AGSIQFHLDH SKMYKFMVND TFSGYPLSES VSRPTQEHES FIGCARELVI 420
421 SGKKLSELCE HNAEVSKKYG KHNATTLWNF IKLFYGSNHF EPPFEHRSSF SNQKNPMNSR 480
481 RATQVASDWP QQRTELGQDL NGNTPETAHE IDVANVDDDT LGSSGVEHPP TSSVILSETQ 540
541 IISEITFDNF ELLRNGFIYV GPTECPKALA FPALHHDVQA ARPQLDLKAS DHEKSPPPHV 600
601 PTVLKVSHVP PIPMWEPHKF VSDALMLMND VGDVQTALTV LIALGEARKL LPIDDALVEH 660
661 WFYTYVDQLH RYELWNEACE VINRSWLRSV QQLNQHSTAM HTNCGECGRP MGGKVGWYCD 720
721 KCKSMQSAKC CVCGLIVRGV YAWCQGCSHG GHIEHLQKYF AKHSKCPKCG HLCAYS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
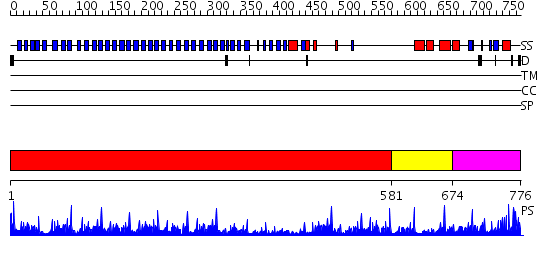
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..580] | 55.39794 | Actin interacting protein 1 | |
| 2 | View Details | [581..673] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [674..776] | 3.11 | Crystal Structure of The Cand1-Cul1-Roc1 Complex |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)