













| General Information: |
|
| Name(s) found: |
XNP-PB /
FBpp0084213
[FlyBase]
XNP-PA / FBpp0084212 [FlyBase] |
| Description(s) found:
Found 36 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1311 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[NAS]
polytene chromosome [IDA] chromosome, centromeric region [IDA] |
| Biological Process: |
transcription
[NAS]
induction of apoptosis [IMP] chromosome organization [IMP] nervous system development [IMP] axon guidance [IMP] glial cell migration [IMP] regulation of JNK cascade [IGI] |
| Molecular Function: |
DNA binding
[IEA]
ATP binding [IEA] DNA helicase activity [NAS] general RNA polymerase II transcription factor activity [NAS] helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGKKNPNARH TDAATPLTTD DSNSSSVSRR ESATESKSAS ESESSPPRSN TKQSRTHKNV 60
61 KASGKATVSS SSDSDQAVAN SSANDEEKEP VCKIRIVPLE KLLASPKTKE RPSRGSQQKN 120
121 VTINDSSDEE PLKGSKLVLP ARKSRNKNAS IIELSDSEEV DEEEESLLVA IPLPKEAQQT 180
181 KPEKNSSKAS KESIEKRQKA QKEATTSSAR AIRSVNGTRR GSLSSERSSR ASSSRAESPP 240
241 RPKRCVVRLK RVSLPKTKPA QKPKKMSSDS EEAATTSKKS RQRRSKSESE ADSDYEAPAA 300
301 EEEEEEERKS SGDEEEAANS SDSEVMPQRK RRRKKSESDK GSSDFEPEEK QKKKGRKRIK 360
361 KTSSGESDGD GDDDKQKNKR KHIRKIIKTK DLDLTTKEAA KEEDDRRKRI EDRQKLYNRI 420
421 FVKSESVEIN ELVLDFDEES KKALLQVDKG LLKKLKPHQV AGVKFMWDAC FETLKESQEK 480
481 PGSGCILAHC MGLGKTLQVV TLSHTLLVNT RRTGVDRVLI ISPLSTVNNW AREFTSWMKF 540
541 ANRNDIEVYD ISRYKDKPTR IFKLNEWFNE GGVCILGYDM YRILANEKAK GLRKKQREQL 600
601 MQALVDPGPD LVVCDEGHLL KNEKTSISKA VTRMRTKRRI VLTGTPLQNN LREYYCMIQF 660
661 VKPNLLGTYK EYMNRFVNPI TNGQYTDSTE RDLRLMKHRS HILHKLLEGC IQRRDYSVLA 720
721 PYLPPKHEYV VYTTLSELQQ KLYGYYMTTH REQSGGDVVG KGARLFQDFQ DLRRIWTHPM 780
781 NLRVNSDNVI AKRLLSNDDS DMEGFICDET DEDEAASNSS DSCETFKSDA SMSGLAASSG 840
841 KVKKRKTRNG NAGGGDSDSD LEMLGGLGGG SSVQKDDPSE WWKPFVEERE LNNVHHSPKL 900
901 LILLRLLQQC EAIGDKLLVF SQSLQSLDVI EHFLSLVDSN TKNYEFEGDV GDFKGCWTSG 960
961 KDYFRLDGSC SVEQREAMCK QFNNITNLRA RLFLISTRAG GLGINLVAAN RVVIFDVSWN1020
1021 PSHDTQSIFR VYRFGQIKPC YIYRLIAMGT MEQKVYERQV AKQATAKRVI DEQQISRHYN1080
1081 QTDLMELYSY ELKPSTEREM PILPKDRLFA EILTEHEKLI FKYHEHDSLL EQEEHENLTE1140
1141 EERKSAWAEY EAEKTRTVQA SQYMSYDRNA FGNQVMGQFG NASGSVTSNK IFGFRSDILL1200
1201 QLLNMKISKD HQELNQNQVI QLVPTYLQQL YNEMNNGDPT MYKDLLNLHS NIVHPSGMYM1260
1261 NPLLYANQNP NAAGYNQGTG GVPPMAGGSV AHGPPAAPAP GFEPDKVYEI D |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
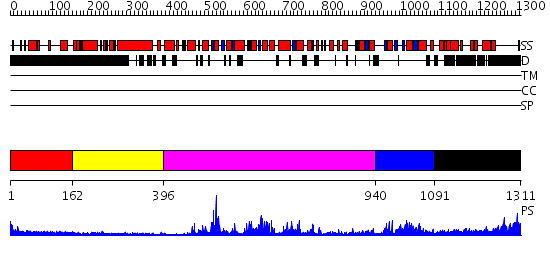
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | 3.417997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [162..395] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [396..939] | 88.0 | Structure of the SWI2/SNF2 chromatin remodeling domain of eukaryotic Rad54 | |
| 4 | View Details | [940..1090] | 29.69897 | Sulfolobus solfataricus SWI2/SNF2 ATPase C-terminal domain | |
| 5 | View Details | [1091..1311] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)