













| General Information: |
|
| Name(s) found: |
loco-PD /
FBpp0083666
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1541 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA]
apical cortex [IDA] plasma membrane [IDA] |
| Biological Process: |
cortical actin cytoskeleton organization
[IMP]
ventral cord development [IMP] septate junction assembly [IMP] dorsal/ventral axis specification, ovarian follicular epithelium [IMP] asymmetric neuroblast division [IMP] establishment of glial blood-brain barrier [IMP] regulation of G-protein coupled receptor protein signaling pathway [IGI][NAS] glial cell differentiation [IMP] cytoplasmic transport, nurse cell to oocyte [IMP] asymmetric protein localization [IMP] |
| Molecular Function: |
G-protein alpha-subunit binding
[IPI]
GTPase activator activity [IDA] receptor signaling protein activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHHHHPPLPI TGASGSTAVG TGAAAAEDAS PAANSGPAPI STSTTPSGSN SQQHQRRRKK 60
61 RANYNYNGIR TVEVRRGYNG FGFTISGQQP CRLSCIISSS PAEQAGLRSG DFLISVNGLN 120
121 VSKLPHETVV QLIGNSFGSI RMQIAENYYS DSSDEENAHA TLRGQLLAAS LRHKPRFLHH 180
181 KAKLHRLRNS PQKKLNPPEA VEPHKSKSSP DHPTLKPVLE DPPLTANLSK AADVANVSAM 240
241 VRAVGSAALE CRVIVGYLGT IEMPKQISHS SKLQTVRSCI RKLRQEKRQP TIVLMCITPD 300
301 SLSLQSSSGG VLATYSSARL NFVSSSSESE NRFFGLVTSA VHNTQIEEEY EPSAGSAAAA 360
361 GHISISHSCH VFVIDTKLIE HAQHLQRAHE FRLQCTRDPI SNLCLEFPNN SEYVVNLVRS 420
421 MYTMRILPPA SRSHQADYEA GAGAAAAGGA VAAAHSPQPS NHSEISTTTS NSDSGIGFNN 480
481 DCTNISDRIL VVDFLAAGAG AAAAAHAGPP TYPAPARPLG IVGIPDNRLT VRAMPDHSAL 540
541 LRSPPASSSL RRPNLLASFN LIQSPATNLT STRSCDDVLN LFVDDSPRTL AAVASMDDIS 600
601 LHSAAPSLDE GHTFAHPACV PRKMRRSLAL ETPTTPHKLS AQVFGQPGSR HSLGFEAIDS 660
661 VQSSVSGACM DQTMDTWASL QNLHKSHKDR TSMSASSSSH CLLEATNSEP DLGNRALPAN 720
721 ASPFRRAWGQ SSFRTPRSDK VAKEQQQLGQ SSPVRRTASM NASDNDMYIK TLMLDSDLKS 780
781 SRSQHQLSLL QVPKVLTTPA PPSAITASVA AEGAAQDHGC PSSWAGSFER MLQDAAGMQT 840
841 FSEFLKKEFS AENIYFWTAC ERYRLLESEA DRVAQAREIF AKHLANNSSD PVNVDSQARS 900
901 LTEEKLADAA PDIFAPAQKQ IFSLMKFDSY QRFIRSDLYK SCVEAEQKNQ PLPYSGLDLD 960
961 ELLKTNFHLG AFSKLKKSAS NAEDRRRKSL LPWHRKTRSK SRDRTEIMAD MQHALMPAPP1020
1021 VPQNAPLTSA SLKLVCGQNS LSDLHSSRSS LSSFDAGTAT GGQGASTESV YSLCRVILTD1080
1081 GATTIVQTRP GETVGELVER LLEKRNLVYP YYDIVFQGST KSIDVQQPSQ ILAGKEVVIE1140
1141 RRVAFKLDLP DPKVISVKSK PKKQLHEVIR PILSKYNYKM EQVQVIMRDT QVPIDLNQPV1200
1201 TMADGQRLRI VMVNSDFQVG GGSSMPPKQS KPMKPLPQGH LDELTNKVFN ELLASKADAA1260
1261 ASEKSRPVDL CSMKSNEAPS ETSSLFERMR RQQRDGGNIP ASKLPKLKKK STSSSQQSEE1320
1321 AATTQAVADP KKPIIAKLKA GVKLQVTERV AEHQDELLEG LKRAQLARLE DQRGTEINFD1380
1381 LPDFLKNKEN LSAAVSKLRK VRASLSPVSK VPATPTEIPQ PAPRLSITRS QQPVSPMKVD1440
1441 QEPETDLPAA TQDQTEFAKA PPPLPPKPKV LPIKPSNWGV AQPTGNYCNK YSPSKQVPTS1500
1501 PKEASKPGTF ASKIPLDLGR KSLEEAGSRC AYLDEPSSSF V |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
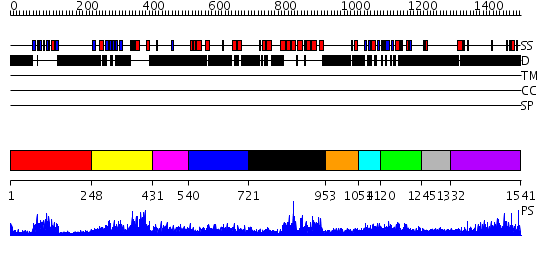
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..247] | 24.69897 | solution structure of the split PH-PDZ Supramodule of alpha-Syntrophin | |
| 2 | View Details | [248..430] | 2.0 | Solution structure of phosphotyrosine interaction domain of mouse Numb protein | |
| 3 | View Details | [431..539] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [540..720] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [721..952] | 59.69897 | Regulator of G-protein signalling 4, RGS4 | |
| 6 | View Details | [953..1053] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1054..1119] | 4.39 | THE RAS-BINDING DOMAIN OF RAF-1 FROM RAT, NMR, 1 STRUCTURE | |
| 8 | View Details | [1120..1244] | 20.30103 | Solution structure of the Ras-binding domain of mouse RGS14 | |
| 9 | View Details | [1245..1331] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1332..1541] | 6.0 | RBP1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 7 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 8 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|