













| General Information: |
|
| Name(s) found: |
cry-PA /
FBpp0083150
[FlyBase]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 542 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] nuclear envelope lumen [IDA] |
| Biological Process: |
DNA repair
[IEA]
UV-A, blue light phototransduction [ISS] gravitaxis [IMP] circadian rhythm [IEP][IGI][IMP][IPI][NAS][TAS] response to blue light [NAS][TAS] regulation of circadian rhythm [IDA][IMP] response to light stimulus [IMP][TAS] phototransduction [IMP] detection of light stimulus involved in magnetoreception [IMP] entrainment of circadian clock [IMP] negative regulation of transcription, DNA-dependent [IMP] locomotor rhythm [IMP] response to magnetism [IMP] protein import into nucleus, translocation [IMP] entrainment of circadian clock by photoperiod [IMP] magnetoreception [IMP] |
| Molecular Function: |
protein binding
[IPI][TAS]
blue light photoreceptor activity [IMP] FAD binding [IDA][IMP] photoreceptor activity [IMP][NAS] [NOT]DNA photolyase activity [IMP][ISS] G-protein coupled photoreceptor activity [IMP][ISS] transcription repressor activity [IMP] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATRGANVIW FRHGLRLHDN PALLAALADK DQGIALIPVF IFDGESAGTK NVGYNRMRFL 60
61 LDSLQDIDDQ LQAATDGRGR LLVFEGEPAY IFRRLHEQVR LHRICIEQDC EPIWNERDES 120
121 IRSLCRELNI DFVEKVSHTL WDPQLVIETN GGIPPLTYQM FLHTVQIIGL PPRPTADARL 180
181 EDATFVELDP EFCRSLKLFE QLPTPEHFNV YGDNMGFLAK INWRGGETQA LLLLDERLKV 240
241 EQHAFERGFY LPNQALPNIH DSPKSMSAHL RFGCLSVRRF YWSVHDLFKN VQLRACVRGV 300
301 QMTGGAHITG QLIWREYFYT MSVNNPNYDR MEGNDICLSI PWAKPNENLL QSWRLGQTGF 360
361 PLIDGAMRQL LAEGWLHHTL RNTVATFLTR GGLWQSWEHG LQHFLKYLLD ADWSVCAGNW 420
421 MWVSSSAFER LLDSSLVTCP VALAKRLDPD GTYIKQYVPE LMNVPKEFVH EPWRMSAEQQ 480
481 EQYECLIGVH YPERIIDLSM AVKRNMLAMK SLRNSLITPP PHCRPSNEEE VRQFFWLADV 540
541 VV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
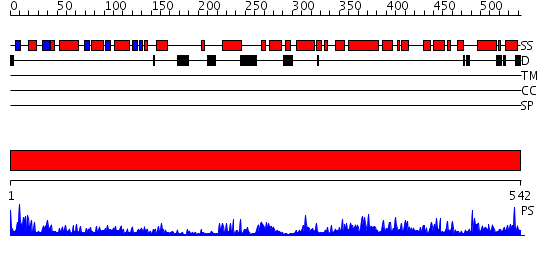
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..542] | 109.0 | Structure of apophotolyase from Anacystis nidulans |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)