













| General Information: |
|
| Name(s) found: |
Fmr1-PA /
FBpp0081675
[FlyBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 684 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lipid particle
[IDA]
micro-ribonucleoprotein complex [TAS] cytoplasm [IDA] RNA-induced silencing complex [IPI][TAS] |
| Biological Process: |
locomotion
[TAS]
regulation of synaptic growth at neuromuscular junction [IMP] germarium-derived oocyte fate determination [IMP] male courtship behavior [TAS] axon guidance [IGI][IMP] [NOT]phototaxis [IMP] larval locomotory behavior [IMP] pole cell formation [IMP] oocyte dorsal/ventral axis specification [IGI] regulation of dendrite morphogenesis [IMP] axonal fasciculation [IMP] mitotic cell cycle, embryonic [IMP] regulation of translation [IMP][NAS] [NOT]chemotaxis [IMP] negative regulation of dendrite morphogenesis [IMP] locomotor rhythm [IMP] negative regulation of translation [IDA][IEP] negative regulation of synaptogenesis [IMP] germ cell development [IMP] regulation of synapse organization [IGI][IMP] mushroom body development [IMP] [NOT]synaptic transmission [IMP] olfactory learning [IMP] sperm axoneme assembly [IMP] brain development [IMP] circadian rhythm [IMP][TAS] cellularization [IDA] neurotransmitter secretion [IMP] regulation of cell proliferation [IGI][IMP] long-term memory [IMP] synapse assembly [NAS] heterochromatin formation [IMP] germ-line cyst formation [IGI][IMP] circadian sleep/wake cycle, sleep [IMP] |
| Molecular Function: |
protein binding
[IPI]
RNA binding [IDA] mRNA binding [IDA][ISS][NAS] protein self-association [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEDLLVEVRL DNGAYYKGQV TAVADDGIFV DVDGVPESMK YPFVNVRLPP EETVEVAAPI 60
61 FEEGMEVEVF TRTNDRETCG WWVGIIKMRK AEIYAVAYIG FETSYTEICE LGRLRAKNSN 120
121 PPITAKTFYQ FTLPVPEELR EEAQKDGIHK EFQRTIDAGV CNYSRDLDAL IVISKFEHTQ 180
181 KRASMLKDMH FRNLSQKVML LKRTEEAARQ LETTKLMSRG NYVEEFRVRD DLMGLAIGSH 240
241 GSNIQAARTV DGVTNIELEE KSCTFKISGE TEESVQRARA MLEYAEEFFQ VPRELVGKVI 300
301 GKNGRIIQEI VDKSGVFRIK VSAIAGDDEQ DQNIPRELAH VPFVFIGTVE SIANAKVLLE 360
361 YHLSHLKEVE QLRQEKMEID QQLRAIQESS MGSTQSFPVT RRSERGYSSD IESVRSMRGG 420
421 GGGQRGRVRG RGGGGPGGGN GLNQRYHNNR RDEDDYNSRG DHQRDQQRGY NDRGGGDNTG 480
481 SYRGGGGGAG GPGNNRRGGI NRRPPRNDQQ NGRDYQHHNH TTEEVRETRE MSSVERADSN 540
541 SSYEGSSRRR RRQKNNNGPS NTNGAVANNN NKPQSAQQPQ QQQPPAPGNK AALNAGDASK 600
601 QNSGNANAAG GASKPKDASR NGDKQQAGTQ QQQPSQVQQQ QAAQQQQPKP RRNKNRSNNH 660
661 TDQPSGQQQL AENVKKEGLV NGTS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
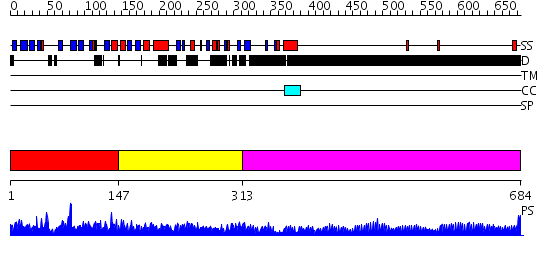
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..146] | 76.30103 | Structure of the N-terminal domain of Fragile X Mental Retardation Protein | |
| 2 | View Details | [147..312] | 30.0 | Solution structure of the N-terminal KH domain of human FXR1 | |
| 3 | View Details | [313..684] | 32.69897 | No description for 1y0fA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)