













| General Information: |
|
| Name(s) found: |
MTA1-like-PA /
FBpp0078382
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 880 amino acids |
Gene Ontology: |
|
| Cellular Component: |
histone deacetylase complex
[IDA]
|
| Biological Process: |
phagocytosis, engulfment
[IMP]
regulation of transcription, DNA-dependent [IEA] |
| Molecular Function: |
transcription factor activity
[IEA]
zinc ion binding [IEA] sequence-specific DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATNMYRVGD YVYVETTPNS PYLIRRIEEL NKNQTGNVEA KVMCFYRRRD LPNPLVQLAD 60
61 KHQLATAEDS PLATKLKKTW LRTPVSEEQA AQAVLDPSIA ALDEERTSPT QTSGGGGSAT 120
121 GNSGSNSSGT SNNNSSSTAI GGGAGGSGGA GGDAEKGEAL TSKQRYQIKH RELFLSRQVE 180
181 SIPATQIRGK CSVTLLNETE SLQSYLNKDD TFFYCLVFDP NQKTLLADKG EIRVGSRYQC 240
241 DIPAKLKDTA TDDRKLEELE SLVWTPEHSL TDRKIDQFLV VSRSIGTFAR ALDCSSSVKQ 300
301 PSLHMSAAAA SRDITLFHAM NILHKHEYSI EESMSSLVPS TGPVLCRDEI EDWSASEANL 360
361 FEEALEKYGK DFNDIRQDFL PWKTLKQIIE YYYMWKTTDR YVQQKRVKAV EAELKLKQVY 420
421 IPQYNNNGKG NGTSTKAGGG IYNGTTNGST DLSSNGKPCE SCGTTKSSQW NSVSSGHSTS 480
481 RLCLSCWEYW RRYGSMKSAT KGDAGEGDAK KKSSSAASTP TATLAGLATT PTAVVDLNDD 540
541 EKISDLTNRQ LHRCSIVNCG KEFKLKTHLA RHYAQAHGIA ISSGSPRPIM KTRTAFYLHT 600
601 NPMTRVARAI CRSIVKPKKA ARQSAYAINA MLVKQEFTNR ISGKSQAEIK KLLLLKPKDR 660
661 GSVTKIANRL GAPGSGPHEW LVLTPKDKMP LPAVVSFPKP PKAPDGSLVY DRVPNKSPDV 720
721 VAVPADKELT IIPTQATSTI RKRAHEDQQL NGTEVTIVPS GPPAKRPNKD PMPSHCPSPE 780
781 QFAAMMAASG QPLSRHHLNG KQKIAQMARG GNGRKQVISW MDAPDDVYFR ANDTHKKTRK 840
841 ILSAVDLRRA ARKPWRTLPI KPAAPEPSSR PIESQIVILD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
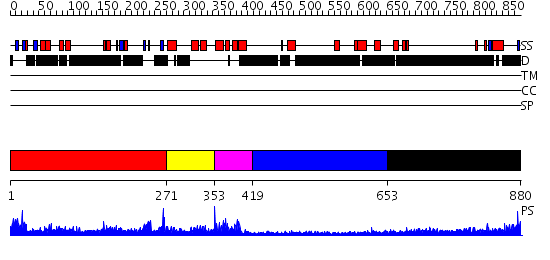
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..270] | 2.0 | Crystal structure of the proximal BAH domain of polybromo | |
| 2 | View Details | [271..352] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [353..418] | 18.522879 | Solution structure of the myb-like DNA-binding domain of mouse MTA3 protein | |
| 4 | View Details | [419..652] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [653..880] | 5.298995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)