













| General Information: |
|
| Name(s) found: |
FBpp0311673
[FlyBase]
FBpp0311672 [FlyBase] UbcD4-PA / FBpp0076124 [FlyBase] |
| Description(s) found:
Found 36 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 199 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
regulation of protein metabolic process
[IEA]
post-translational protein modification [IEA] |
| Molecular Function: |
ubiquitin-protein ligase activity
[NAS]
small conjugating protein ligase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MANMAVSRIK REFKEVMRSE EIVQCSIKIE LVNDSWTELR GEIAGPPDTP YEGGKFVLEI 60
61 KVPETYPFNP PKVRFITRIW HPNISSVTGA ICLDILKDNW AAAMTLRTVL LSLQALLAAA 120
121 EPDDPQDAVV AYQFKDKYDL FLLTAKHWTN AYAGGPHTFP DCDSKIQRLR DMGIDEHEAR 180
181 AVLSKENWNL EKATEGLFS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
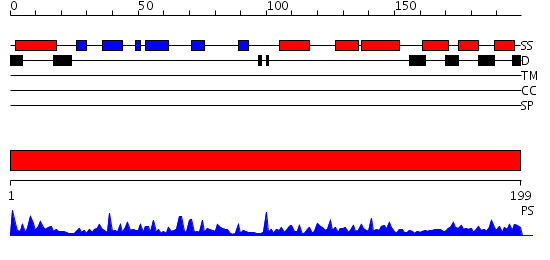
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..199] | 54.69897 | Ubiquitin-conjugating enzyme E2-25 kDa (Huntington interacting protein 2) |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)