













| General Information: |
|
| Name(s) found: |
pain-PA /
FBpp0072323
[FlyBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 913 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA]
integral to membrane [NAS] |
| Biological Process: |
response to heat
[TAS]
ion transport [IEA] regulation of female receptivity [IMP] transmembrane transport [IEA] [NOT]thermotaxis [IMP] calcium ion transport [ISS] sensory perception of pain [IMP][NAS] feeding behavior [IMP] copulation [IMP] response to mechanical stimulus [NAS] detection of mechanical stimulus involved in sensory perception [TAS] |
| Molecular Function: |
ion channel activity
[IEA][TAS]
calcium channel activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDFNNCGFID PQAQLAGALA KQDIRQFVAA LDSGALADLQ DDRHTSIYEK ALSTPGCRDF 60
61 IEACIDHGSQ VNYINKKLDK AAISYAADSR DPGNLAALLK YRPGNKVQVD RKYGQLTPLN 120
121 SLAKNLTDEN APDVYSCMQL LLDYGASPNI VDQGEFTPLH HVLRKSKVKA GKKELIQLFL 180
181 DHPELDIDSY RNGEVRRLLQ AQFPELKLPE ERHTGPEIDI QTLQRTLRDG DETLFEQQFA 240
241 EYLQNLKGGA DNQLNAHQEE YFGLLQESIK RGRQRAFDVI LSTGMDINSR PGRANEANLV 300
301 ETAVIYGNWQ ALERLLKEPN LRLTPDSKLL NAVIGRLDEP PYDGSSHQRC FELLINSDRV 360
361 DINEADSGRL VPLFFAVKYR NTSAMQKLLK NGAYIGSKSA FGTLPIKDMP PEVLEEHFDS 420
421 CITTNGERPG DQNFEIIIDY KNLMRQERDS GLNQLQDEMA PIAFIAESKE MRHLLQHPLI 480
481 SSFLFLKWHR LSVIFYLNFL IYSLFTASII TYTLLKFHES DQRALTAFFG LLSWLGISYL 540
541 ILRECIQWIM SPVRYFWSIT NIMEVALITL SIFTCMESSF DKETQRVLAV FTILLVSMEF 600
601 CLLVGSLPVL SISTHMLMLR EVSNSFLKSF TLYSIFVLTF SLCFYILFGK SVEEDQSKSA 660
661 TPCPPLGKKE GKDEEQGFNT FTKPIEAVIK TIVMLTGEFD AGSIQFTSIY TYLIFLLFVI 720
721 FMTIVLFNLL NGLAVSDTQV IKAQAELNGA ICRTNVLSRY EQVLTGHGRA GFLLGNHLFR 780
781 SICQRLMNIY PNYLSLRQIS VLPNDGNKVL IPMSDPFEMR TLKKASFQQL PLSAAVPQKK 840
841 LLDPPLRLLP CCCSLLTGKC SQMSGRVVKR ALEVIDQKNA AEQRRKQEQI NDSRLKLIEY 900
901 KLEQLIQLVQ DRK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
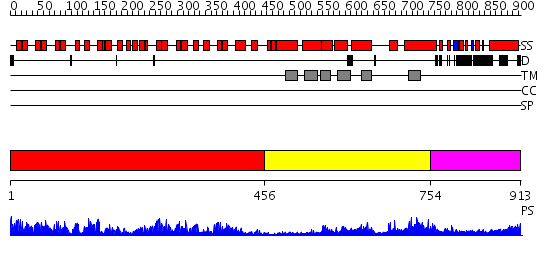
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..455] | 91.69897 | Ankyrin-R | |
| 2 | View Details | [456..753] | 17.30103 | Mammalian Shaker Kv1.2 potassium channel- beta subunit complex | |
| 3 | View Details | [754..913] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|