













| General Information: |
|
| Name(s) found: |
CG5594-PB /
FBpp0072047
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1028 amino acids |
Gene Ontology: |
|
| Cellular Component: |
integral to membrane
[ISS]
|
| Biological Process: |
mechanosensory behavior
[IMP]
sodium ion transport [IEA] transmembrane transport [IEA] transport [ISS] chloride transport [IEA] amino acid transport [ISS] response to mechanical stimulus [IMP] |
| Molecular Function: |
potassium:chloride symporter activity
[ISS]
amino acid transmembrane transporter activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQSNSAKDDS DEMEIIPSEH NGTVHETKNE DSGDIHRAEE NQKSSIDPNL YLYDDDLETR 60
61 PHISTFISSI ANYENTIPAA TDPDAKPAAP SARMGTLIGV FLPCIQNIFG VILFIRLTWV 120
121 VGTAGAVCGF LIVLTCCCVT MLTAISMSAI ATNGVVPAGG SYFMISRSLG PEFGGAVGML 180
181 FYTGTTLAAA MYIVGAVEIV LTYMAPWASI FGDFTKDADA MYNNFRVYGT LLLIFMGLIV 240
241 FLGVKFVNKF ATVALACVIL SIIAVYVGIF DNIHGNEKLY MCVLGKRLLK DIPLENCTKE 300
301 DSFLRDIYCP DGKCEEYYLA NNVTKVKGIK GLASGVFYDN IFPSFLEKGQ FISYGKSAID 360
361 IENTSGESYN QIMADITTSF TLLIGIFFPS VTGIMAGSNR SGDLADAQKS IPIGTICAIL 420
421 TTSTVYLSSV MFFAGTVDNL LLRDKFGQSI GGKLVVANIA WPNQWVILIG SFLSTLGAGL 480
481 QSLTGAPRLL QAIARDEIIP FLAPFAKSSK RGEPTRALLL TIVICQCGIL LGNVDLLAPL 540
541 LSMFFLMCYG FVNLACAVQT LLRTPNWRPR FKFYHWSLSL IGLTLCISVM IMTSWYFALI 600
601 AMGMAIIIYK YIEYRGAEKE WGDGIRGMAL TAARYSLLRL EEGPPHTKNW RPQILVLSKL 660
661 NDNLLPKYRK IFSFATQLKA GKGLTICVSV IKGDHTKITN KAVDAKATLR KYMTDEKVKG 720
721 FCDVLVAQQI GEGLSSVIQT IGLGGMKPNT VIIGWPYSWR QEGRNSWKTF IQTVRTVAAC 780
781 HMALMVPKGI NFYPESNHKI GGNIDIWWIV HDGGLLMLLP FLLKQHRTWR NCKLRIFTVA 840
841 QIEDNSIQMK KDLKTFLYHL RIEADVEVVE MNNSDISAYT YERTLMMEQR NQMLRALGLN 900
901 KKENSKVDSQ NDEKRNSIDL DGPENADTPE TTSNKDESTE KADGDFKSSV KPDEFNVRRM 960
961 HTAIKLNEVI VEKSQDAQLV IMNLPGPPRE VRAERERNYM EFLEVLTEGL EKVLMVRGGG1020
1021 REVITIYS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
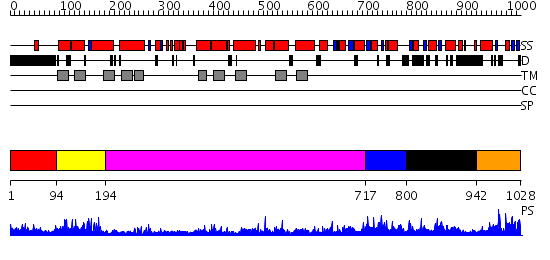
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..93] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [94..193] | 4.522879 | Crystal structure of LEUTAA, a bacterial homolog of Na+/Cl--dependent neurotransmitter transporters | |
| 3 | View Details | [194..716] | 6.045757 | Bacterial aa3 type cytochrome c oxidase subunit I | |
| 4 | View Details | [717..799] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [800..941] | 1.130966 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [942..1028] | 1.080994 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|