













| General Information: |
|
| Name(s) found: |
CG5602-PA /
FBpp0072041
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 747 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
DNA repair
[IEA]
DNA replication [IEA] DNA recombination [IEA] |
| Molecular Function: |
DNA binding
[IEA]
ATP binding [IEA] DNA ligase (ATP) activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQKSITSFFK KKSDATDSPS PPKKVPKIDA KTELPDEPHI KSESASPETK PKVEPMSVDS 60
61 EEKTSPVKNV KKEPKEVDDK TTDKKVTTIG LNSTAATKED VENYDPSADS YHPLKNAYWK 120
121 DKKVTPYLAL ARTFQVIEET KGRLKMIDTL SNFFCSVMLV SPEDLVPSVY LSINQLAPAY 180
181 EGLELGVAET TLMKAICKAT GRNLAHIKSQ TQLTGDLGIV AEQSRVSQRM MFQPAPLNVR 240
241 DVFRKLREIA KLSGQSKMDL VYNMFVACRS SEARFFIRSL IGKLRIGIAE QSLLTALAIG 300
301 LVKKNHIDDC KASKVPDVYK DEIVDTTLLL KTAYCQCPNY DIIIPAILKY DIKELQERCP 360
361 MHPGMPLRPM LAQPTKGVHE VFERFGGMQI TCEWKYDGER AQIHRNEKGE ISIFSRNSEN 420
421 NTAKYPDLIA RSTALLKGDV KSYIIDSEIV AWDVERKQIL PFQVLSTRKR KNVDIEEIKV 480
481 QVCVYIFDLL YINGTALVTK NLSERRKLLL EHFQEVEGEW KFATALDTND IDEVQQFLEE 540
541 SIKGNCEGLM VKTLDEEATY EIAKRSRNWL KLKKDYLSNV GDSLDLVVIG GYKGKGRRTG 600
601 TYGGFLLACY DTENEEYQSI CKIGTGFTDE DLQTHSEFLG KHVTSAAKSY YRYDPSLEPD 660
661 HWFEPVQVWE VKCADLSLSP IHRAAIGIVD GERGISLRFP RFIRIRDDKN SENATDANQV 720
721 AHMYQSQDQV KNNQKSSTQM EMEDEFY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
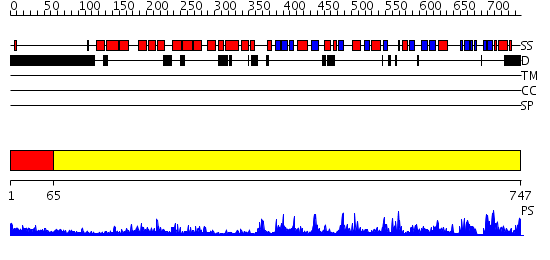
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..64] | 4.09691 | Solution structure of the zinc-finger domain from DNA ligase IIIa | |
| 2 | View Details | [65..747] | 177.0 | Crystal Structure of Human DNA Ligase I bound to 5'-adenylated, nicked DNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)