













| General Information: |
|
| Name(s) found: |
apt-PB /
FBpp0071975
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 489 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
negative regulation of oskar mRNA translation
[NAS][TAS]
heart development [TAS] central nervous system development [IMP] primary branching, open tracheal system [TAS] neuromuscular synaptic transmission [IMP] positive regulation of transcription from RNA polymerase II promoter [IDA] regulation of transcription from RNA polymerase II promoter [NAS] open tracheal system development [IMP][TAS] |
| Molecular Function: |
transcription factor activity
[IDA]
protein binding [IPI] RNA polymerase II transcription factor activity [NAS] mRNA binding [TAS] sequence-specific DNA binding [IDA] transcription activator activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTMANGAVNK QYSATDLEAF MKIAANWQNS NHNLLEGVDI KTEMTQYMNS PSMNMYKKRE 60
61 RTQNWNHQEK KYLLDLCRKD MRIIENKRLD AGLTAVKNKA WKIIHQKFSN QFGTDRTCNR 120
121 LKEQWRRMKA CTRNEILDYN NRLARFGAEV ADRKKPSPFT FEVWDFMQEA KKACKSEALD 180
181 GIDYSKIPLA LEEGFEYRED YKFNPEDHDM DDSRTPPQEL CDVDIKEEEN NETFNETFNH 240
241 HHLNQSDILG SGERLSVSPN HSISRNGIHE AESPATISTS NVLDSSGAYM AGGGDMSGFQ 300
301 ANFNMNNISA TLEALNALRS GQFPSAAAAA AAAIHQHQAQ AQVAMHHQHN NNNNNNNNNE 360
361 DDEDSMKPQS KRQRTSSGSM LGSEDQAPVL APSVPASSDS QALERSGSSN GLTMPGTAVS 420
421 TNANSSSSNN NNFELRMFME MQSKEHMMRM KILEVQLQAA KHSRDLVEIN KTLALQKLQL 480
481 ELASKRLPS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
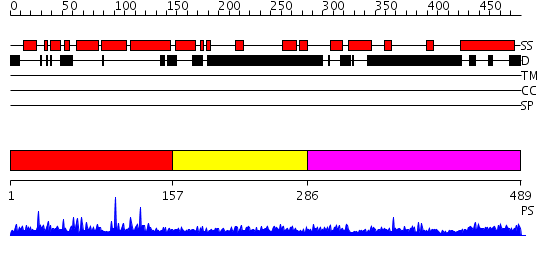
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..156] | 23.065997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [157..285] | 1.042986 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [286..489] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)