













| General Information: |
|
| Name(s) found: |
FBpp0302010
[FlyBase]
CG9485-PE / FBpp0289624 [FlyBase] CG9485-PA / FBpp0071525 [FlyBase] |
| Description(s) found:
Found 36 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1542 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
glycogen biosynthetic process
[IEA]
|
| Molecular Function: |
cation binding
[IEA]
amylo-alpha-1,6-glucosidase activity [ISS] 4-alpha-glucanotransferase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTMGKETES HSIPISEGQD AEHILYRLKR GSKLSVHPDA SLLGRKIVLY TNYPAEGQKF 60
61 VRTEYRVLGW QLSNGKQITS VMHPEAHVVD TDIRSQVELN MSGTYHFYFR YLERPDTGCS 120
121 GADGALYVQV EPTLHVGPPG AQKTIPLDSV RCQTVLAKLL GPLDTWEPKL RVAKEAGYNV 180
181 IHFTPIQELG GSRSCYSLRD QLKVNSHFAP QKGGKISFED VEKVIKKCRQ EWGVASICDI 240
241 VLNHTANESD WLLQHPDATY SCATCPYLRP AFLLDATFAQ CGADIAEGSL EHVGVPAVIE 300
301 QECHLEALKY QLHTSYMSKV NIHELYQCDV MKYVNEFMSQ VRTREPPKNV ANECRFQEIQ 360
361 LIQDPQYRRL ASTINFELAL EIFNAFHGDC FDEESRFRKC AETLRRHLDA LNDRVRCEVQ 420
421 GYINYAIDNV LAGVRYERVQ GDGPRVKEIS EKHSVFMVYF THTGTQGKSL TEIEADMYTK 480
481 AGEFFMAHNG WVMGYSDPLR DFAEEQPGRA NVYLKRELIS WGDSVKLRFG RRPEDSPYLW 540
541 QHMTEYVQTT ARIFDGVRLD NCHSTPLHVA EYLLDAARKI NPELYVVAEL FTNSDYTDNV 600
601 FVNRLGITSL IREALSAWDS HEQGRLVYRY GGVPVGGFQA NSSRHEATSV AHALFLDLTH 660
661 DNPSPVEKRS VYDLLPSAAL VSMACCATGS NRGYDELVPH HIHVVDEERT YQEWGKGVDS 720
721 KSGIMGAKRA LNLLHGQLAE EGFSQVYVDQ MDPNVVAVTR HSPITHQSVI LVAHTAFGYP 780
781 SPNAGPTGIR PLRFEGVLDE IILEASLTMQ SDKPFDRPAP FKKDPNVING FTQFQLNLQE 840
841 HIPLAKSTVF QTQAYSDGNN TELNFANLRP GTVVAIRVSM HPGPRTSFDK LQKISAALRI 900
901 GSGEEYSQLQ AIVSKLDLVA LSGALFSCDD EERDLGKGGT AYDIPNFGKI VYCGLQGFIS 960
961 LLTEISPKND LGHPLCNNLR DGNWMMDYIS DRLTSYEDLK PLSAWFKATF EPLKNIPRYL1020
1021 IPCYFDAIVS GVYNVLINQV NELMPDFIKN GHSFPQSLAL STLQFLSVCK SANLPGFSPA1080
1081 LSPPKPPKQC VTLSAGLPHF STGYMRCWGR DTFIALRGSM FLTGRYNEAR FIIIGFGQTL1140
1141 RHGLIPNLLD SGSKPRFNCR DAIWWWMYCI KQYVEDAPKG AEILKDKVSR IFPYDDADAH1200
1201 APGAFDQLLF DVMQEALQVH FQGLQYRERN AGYEIDAHMV DQGFNNQIGI HPETGFVFGG1260
1261 NNFNCGTWMD KMGSSQKAGN KGRPSTPRDG SAVELVGLQY AVLRFMQSLA EKEVIPYTGV1320
1321 ERKGPSGEVT KWSYKEWADR IKNNFDKYFF VSESETCSVA NKKLIYKDSY GATQSWTDYQ1380
1381 LRCNFPITLT VAPDLCNPQN AWRALERAKK YLLGPLGMKT MDPEDWNYRA NYDNSNDSTD1440
1441 CTVAHGANYH QGPEWVWPIG FYLRARLIFA KKCGHLDETI AETWAILRAH LRELQTSHWR1500
1501 GLPELTNDNG SYCGDSCRTQ AWSVAAILEV LYDLHSLGAD VA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
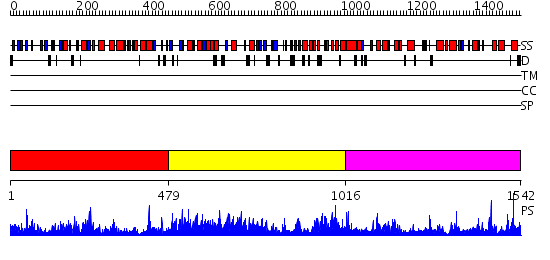
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..478] | 77.0 | Neopullulanase, central domain; Neopullulanase | |
| 2 | View Details | [479..1015] | 3.37 | COMPLEX OF A (D229N/E257Q) DOUBLE MUTANT CGTASE FROM BACILLUS CIRCULANS STRAIN 251 WITH MALTOTETRAOSE AT 120 K AND PH 9.1 OBTAINED AFTER SOAKING THE CRYSTAL WITH ALPHA-CYCLODEXTRIN | |
| 3 | View Details | [1016..1542] | 77.522879 | No description for 2jf4A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)