













| General Information: |
|
| Name(s) found: |
unc-104-PC /
FBpp0112001
[FlyBase]
unc-104-PB / FBpp0112000 [FlyBase] |
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1670 amino acids |
Gene Ontology: |
|
| Cellular Component: |
kinesin complex
[ISS]
microtubule associated complex [IMP] |
| Biological Process: |
neuromuscular junction development
[IMP]
vesicle-mediated transport [IMP] synaptic vesicle transport [IMP] vesicle transport along microtubule [IMP] larval locomotory behavior [IMP] synaptic vesicle maturation [IMP] anterograde synaptic vesicle transport [IMP] |
| Molecular Function: |
ATP binding
[IEA]
microtubule motor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSVKVAVRV RPFNSREIAR ESKCIIEMAG ATTAITNPKV PPNTSDSVKR FNFDYSYWSH 60
61 DHHDADFSTQ SMVYKDIGEE MLQHSFDGYN VCIFAYGQTG AGKSYTMMGR QEEQQEGIIP 120
121 MICKDLFTRI QDTETDDLKY SVEVSYMEIY CERVRDLLNP KNKGNLRVRE HPLLGPYVED 180
181 LSKLAVTDYQ DIHDLIDEGN KARTVAATNM NETSSRSHAV FTIFFTQRRH DLMTNLTTEK 240
241 VSKISLVDLA GSERADSTGA KGTRLKEGAN INKSLTTLGK VISALAEVAS KKKNTKKADF 300
301 IPYRDSALTW LLRENLGGNS KTAMIAAISP ADINYDETLS TLRYADRAKQ IVCKAVVNED 360
361 ANAKLIRELK EEIQKLRDLL KAEGIEVQEE DELTKSTVIK SPTKSRNRNG STTEMAVDQL 420
421 QASEKLIAEL NETWEEKLKR TEEIRVQREA VFAEMGVAVK EDGITVGVFS PKKTPHLVNL 480
481 NEDPNLSECL LYYIKEGLTR LGTHEANVPQ DIQLSGSHIL KEHCTFENKN STVTLLPHKD 540
541 AIIYVNGRKL VEPEVLKTGS RVILGKNHVF RFTNPEQARE LRDKIETENE AENEVEKTDT 600
601 QQVDWNFAQC ELLEKQGIDL KAEMKKRLDN LEEQYKREKL QADQQFEEQR KTYEARIDAL 660
661 QKQVEEQSMT MSMYSSYSPE DFHQEEDVYT NPMYESCWTA REAGLAAWAF RKWRYHQFTS 720
721 LRDDLWGNAI FLKEANAISV ELKKKVQFQF TLLTDTLYSP LPPELASTVA PVHQEDEFGA 780
781 PPVSKTLVAV EVTDTKNGAT HHWSLEKLRQ RLELMREMYH NEAEMSPTSP DYNVESLTGG 840
841 DPFYDRFPWF RMVGRSFIYL SNLLYPVPLV HKVAIVNERG DVRGYLRIAV QPVLDEESID 900
901 FNNGVKQSAR LVFNEDDAKP KYRALNEKDD VQRYIDNGGL DSKLEELEDV DSGRGIDSNS 960
961 ASECHENSEE PGEHLQVGKE FTFRVTVLQA TGIGAEYADI FCQFNFLHRH EEAFSTEPVK1020
1021 NSASGAPLGF YHVQNITVPV TKSFIEYLKT QPIMFKIFGH YQTHPLHKDA KQDFVSRPPP1080
1081 RRMLPPSIPI SQPVRSPKFG PLPCAPTSTV LAKHDVLVWF EICELAPNGE YVPSVVEHSD1140
1141 DLPCRGLFLL HQGIQRRIRI TIVHEPTTEV KWKDINELVV GRIRNTPESS DEQDEDACVL1200
1201 SLGLFPGEAL EVPGDDRSFY RFEAAWDSSL HNSALLNRVS QGGETIYITL SAYLELENCA1260
1261 RPAIITKDLS MVIYGRDART GPRSLKHLFS GQYRNPEANR LTGVYELALR RASEAGSPGV1320
1321 QRRQRRVLDT SSTYVRGEEN LHGWRPRGDS LIFDHQWELE KLTRLEEVGR MRHLLLLRER1380
1381 LGMDTNPNPT TKTEKDVCNL AARAATSPVH MVIPQSPQTP VKDPQQIIPE REYNQREQDL1440
1441 MLKCLKLVQG RYTKSEANDT QTQSDVSPSD EGCADMTVSC ISSNSMENNK FVIRRRLCSP1500
1501 DRADAPNGWE APAPATQPAL PLRLYVPELE EIRVSPVVAR KGLLNVLEHG GSGWKKRWVI1560
1561 VRRPYVFIYR SEKDPVERAV LNLATAHVEC SEDQAAMVKI PNTFSVVTKH RGYLLQTLGD1620
1621 KEVHDWLYAI NPLLAGQIKS RLARRTLEPA SQTASQIQAT NAANANSASK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
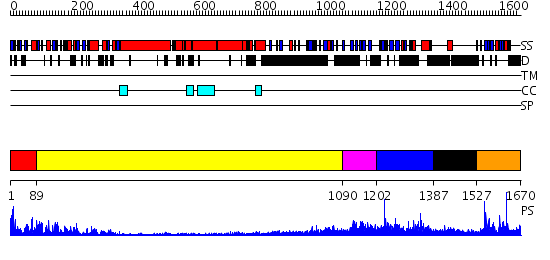
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..88] | 94.39794 | Kinesin | |
| 2 | View Details | [89..1089] | 14.0 | Heavy meromyosin subfragment | |
| 3 | View Details | [1090..1201] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [1202..1386] | 1.061973 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [1387..1526] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1527..1670] | 27.30103 | Tapp1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)