













| General Information: |
|
| Name(s) found: |
CG7813-PA /
FBpp0086272
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 734 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)
[IEA]
mitochondrial proton-transporting ATP synthase complex [ISS] |
| Biological Process: |
ATP synthesis coupled proton transport
[IEA]
|
| Molecular Function: |
hydrogen-exporting ATPase activity, phosphorylative mechanism
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MWEKLVNCIK SNGGNGGVKN VSCQVVELAD LMKQVPPNQM HKFKMFAKKH EEYKDRVRKY 60
61 PESMPTIDWE YYRQNVREEF VDWVKGYETK YDKLHSVFEN RHAIVDHKRY FELVDEEKKV 120
121 VTKCISEYKA ESDKRIQELT EKLEFVKAMR PYSEMTMEEF CFARPHLAPD FINKPTFWPH 180
181 TPEEQMPGPS DPEAAAALHH EEEPEPPKKP APEKLPDKPS GEGKPPAAAA PKEPKEPVVD 240
241 TSQLAEKATL IAKDLMAKAI VLFNSLKEKM SGLAKNVQKK ADAAKAARSE TASKSATPTA 300
301 APTKSLDSIT ERESGPNICN QTIIRSEEAE ANPEVKARHT NLSIEADPCD AQEERAREIA 360
361 RTLERKRQLK EAEEWEKHKT KKDPCDAEKE TDPCKLMEEE TVCKPDPCKA KEEEAVCKPD 420
421 PCQQKEKEDD ICKPDPCKPK DEEGSCEDEE DPCKKKPKCD GDYQFGGDNE GGSTKSKQQD 480
481 QVFINISECS KELAKQEEKK SGDSGKPKEK ASLSEIPQGS DQKPVLGMQI TESKDSKKDH 540
541 KASIEGANEI GPVYTDPNQL AELLVQKKEK PVIIEEAQKP EDDKPITIYP KLEISANPKG 600
601 DQVKQEAEGQ KSPKDMAKQV FTMASGAATL LTEATNTLED LKRKKEARLE ALEQAYTSAQ 660
661 RQAQGALAEA SKAVEAANNL AIRSAEQTGE VSSRDREALE MAEKHAILAK MLAGRAVALK 720
721 DEIARVLKDL KKKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
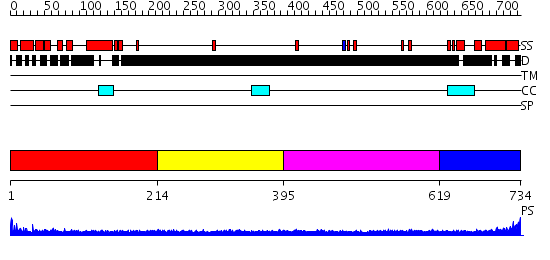
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..213] | 4.83 | No description for 2clyB was found. | |
| 2 | View Details | [214..394] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [395..618] | 1.200991 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [619..734] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)