













| General Information: |
|
| Name(s) found: |
FBpp0305367
[FlyBase]
ttv-PA / FBpp0086624 [FlyBase] |
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 760 amino acids |
Gene Ontology: |
|
| Cellular Component: |
integral to membrane
[NAS]
endoplasmic reticulum [IDA] Golgi apparatus [IDA] intrinsic to endoplasmic reticulum membrane [IEA] |
| Biological Process: |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process
[IMP][NAS]
heparan sulfate proteoglycan biosynthetic process [IDA][IMP][NAS][TAS] glycosaminoglycan biosynthetic process [IMP] smoothened signaling pathway [IMP][NAS] Wnt receptor signaling pathway [IGI][IMP] N-acetylglucosamine metabolic process [ISS] decapentaplegic receptor signaling pathway [IMP] heparin biosynthetic process [IMP] germ cell migration [IMP] regulation of smoothened signaling pathway [IMP] |
| Molecular Function: |
protein binding
[IPI]
acetylglucosaminyltransferase activity [ISS][NAS] glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity [ISS] glucuronosyltransferase activity [NAS] N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQAKKRYILV FVSCAFLAYA YFGGYRLKVS PLRPRRAQHE SAKDGGVQPH EQLPSFLGAH 60
61 DMQELQLLQS NQSKSLDSSK HLVTRKPDCR METCFDFTRC YDRFLVYIYP PEPLNSLGAA 120
121 PPTSANYQKI LTAIQESRYY TSDPTAACLF VLGIDTLDRD SLSEDYVRNV PSRLARLPYW 180
181 NNGRNHIIFN LYSGTWPDYA ENSLGFDAGE AILAKASMGV LQLRHGFDVS IPLFHKQFPL 240
241 RAGATGTVQS NNFPANKKYL LAFKGKRYVH GIGSETRNSL FHLHNGRDMV LVTTCRHGKS 300
301 WRELQDNRCD EDNREYDRYD YETLLQNSTF CLVPRGRRLG SFRFLEALQA GCIPVLLSNA 360
361 WVLPFESKID WKQAAIWADE RLLLQVPDIV RSIPAERIFA LRQQTQVLWE RYFGSIEKIV 420
421 FTTFEIIRER LPDYPVRSSL VWNSSPGALL TLPTFADSSR YMPFLLNSMG AEPRHNYTAV 480
481 IYVQIGAALG PNAALYKLVR TITKSQFVER ILVLWAADRP LPLKKRWPPT SHIPLHVISL 540
541 GGSTRSQGAG PTSQTTEGRP SISQRFLPYD EIQTDAVLSL DEDAILNTDE LDFAYTVWRD 600
601 FPERIVGYPA RAHFWDDSKN AWGYTSKWTN YYSIVLTGAA FYHRYYNYLY TNWLSLLLLK 660
661 TVQQSSNCED ILMNLLVSHV TRKPPIKVTQ RKGYKDRETG RSPWNDPDHF IQRQSCLNTF 720
721 AAVFGYMPLI RSNLRMDPML YRDPVSNLRK KYRQIELVGS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
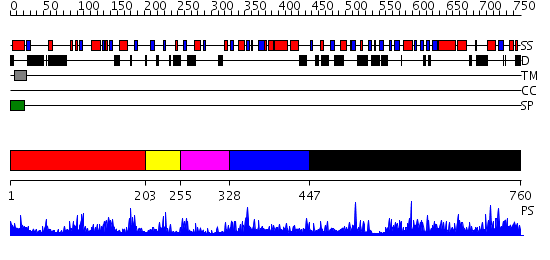
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..202] | 3.111977 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [203..254] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [255..327] | 2.126991 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [328..446] | 7.128991 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [447..760] | 89.39794 | Alpha-1,4-N-acetylhexosaminyltransferase (Alpha-GalNAcT EXTL2) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|