













| General Information: |
|
| Name(s) found: |
FBpp0294063
[FlyBase]
Tfb1-PA / FBpp0086678 [FlyBase] Tfb1-PC / FBpp0086677 [FlyBase] Tfb1-PB / FBpp0086676 [FlyBase] |
| Description(s) found:
Found 56 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 585 amino acids |
Gene Ontology: |
|
| Cellular Component: |
holo TFIIH complex
[ISS]
|
| Biological Process: |
transcription initiation from RNA polymerase II promoter
[ISS]
|
| Molecular Function: |
general RNA polymerase II transcription factor activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTSSEDVLL QMGEVRYKKG DGTLYVMNER VAWMAEHRDT VTVSHRYADI KTQKISPEGK 60
61 PKVQLQVVLH DGNTSTFHFV NRQGQAAMLA DRDKVKELLQ QLLPNFKRKV DKDLEDKNRI 120
121 LVENPNLLQL YKDLVITKVL TSDEFWATHA KDHALKKMGR SQEIGVSGAF LADIKPQTDG 180
181 CNGLKYNLTS DVIHCIFKTY PAVKRKHFEN VPAKMSEAEF WTKFFQSHYF HRDRLTAGTK 240
241 DIFTECGKID DQALKAAVQQ GAGDPLLDLK KFEDVPLEEG FGSVAGDRNV VNSGNIVHQN 300
301 MIKRFNQHSI MVLKTCANVT SAPSTMTNGT NNANGPVSQS AYTNGMNGKG QATATATKSS 360
361 SDQVDKDEPQ SKKQRLMEKI HYVDLGDPIL EGDDSANGEK AKSKHFELSK VERYLNGPVQ 420
421 NSMYDNHNDP MSLEEVQYKL VRNSESWLNR NVQRTFICSK AAVNALGELS PGGSMMRGFQ 480
481 EQSAGQLVPN DFQRELRHLY LSLSELLKHF WSCFPPTSEE LETKLQRMHE TLQRFKMAKL 540
541 VPFENRAMHE LSPLRSSLTQ HLNQLLRTAN SKFATWKERK LRNTR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
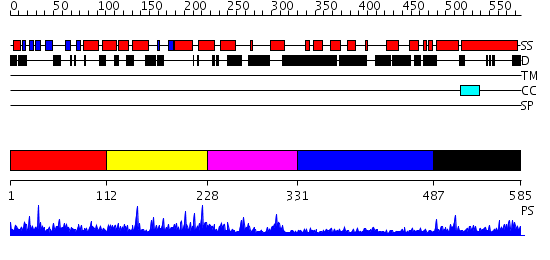
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..111] | 37.30103 | Solution structure of the N-terminal PH/PTB domain of the TFIIH P62 subunit | |
| 2 | View Details | [112..227] | 1.68 | Solution structure of the BSD domain of human Synapse associated protein 1 | |
| 3 | View Details | [228..330] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [331..486] | 1.037986 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [487..585] | 2.036988 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)