













| General Information: |
|
| Name(s) found: |
FBpp0308978
[FlyBase]
mam-PB / FBpp0086723 [FlyBase] mam-PA / FBpp0086722 [FlyBase] |
| Description(s) found:
Found 40 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1594 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA][ISS][NAS]
nuclear speck [IEA] |
| Biological Process: |
nervous system development
[NAS]
imaginal disc-derived wing morphogenesis [IMP] asymmetric cell division [IMP] imaginal disc-derived wing margin morphogenesis [IMP] ectoderm development [TAS] apposition of dorsal and ventral imaginal disc-derived wing surfaces [IMP] positive regulation of transcription from RNA polymerase II promoter [IEA] compound eye development [TAS] mesodermal cell fate determination [IMP] bristle morphogenesis [IMP] heart morphogenesis [IMP] Notch signaling pathway [IEA][NAS] germ-line stem cell maintenance [IMP] somatic stem cell maintenance [IMP] |
| Molecular Function: |
transcription coactivator activity
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDAGGLPVFQ SASQAAAVAQ QQQQQQQQQQ QHLNLQLHQQ HLGLHLQQQQ QLQLQQQQHN 60
61 AQAQQQQIQV QQQQQQQQQQ QQQQHSPYNA NLGATGGIAG ITGGNGAGGP TNPGAVPTAP 120
121 GDTMPTKRMP VVDRLRRRME NYRRRQTDCV PRYEQAFNTV CEQQNQETTV LQKRFLESKN 180
181 KRAAKKTDKK LPDPSQQHQQ QQHQQQQQHQ QHQQHQQAQT MLAGQLQSSV HVQQKFLKRP 240
241 AEDVDNGPDS FEPPHKLPNN NNNSNSNNNN GNANANNGGN GSNTGNNTNN NGNSTNNNGG 300
301 SNNNGSENLT KFSVEIVQQL EFTTSAANSQ PQQISTNVTV KALTNTSVKS EPGVGGGGGG 360
361 GGGGGSGNNN NNGGGGGGGN GNNNNNGGDH HQQQHQQQQQ QQGGGLGGLG NNGRGGGPGG 420
421 MATGPGGVAG GLGGMGMPPN MMSAQQKSAL GNLANLVECK REPDHDFPDL GSLDKDGGGG 480
481 QFPGFPDLLG DDNSENNDTF KDLINNLQDF NPSFLDGFDE KPLLDIKTED GIKVEPPNAQ 540
541 DLINSLNVKS EGGLGHGFGG FGLGLDNPGM KMRGGNPGNQ GGFPNGPNGG TGGAPNAGGN 600
601 GGNSGNLMSE HPLAAQTLKQ MAEQHQHKNA MGGMGGFPRP PHGMNPQQQQ QQQQQQQQQQ 660
661 AQQQHGQMMG QGQPGRYNDY GGGFPNDFGL GPNGPQQQQQ AQQQQPQQQH LPPQFHQQKG 720
721 PGPGAGMNVQ QNFLDIKQEL FYSSQNDFDL KRLQQQQAMQ QQQQQQHHQQ QQQQQQPKMG 780
781 GVPNFNKQQQ QQQVPQQQLQ QQQQQQQQQQ QQQQQQYSPF SNQNPNAAAN FLNCPPRGGP 840
841 NGNQQPGNLA QQQQQPGAGP QQQQQRGNAA NGQQNNPNAG PGGNTPNAPQ QQQQQSTTTT 900
901 LQMKQTQQLH ISQQGGGAHG IQVSAGQHLH LSGDMKSNVS VAAQQGVFFS QQQAQQQQQQ 960
961 QQPGGTNGPN PQQQQQQPHG GNAGGGVGVG VGVGVGNGGP NPGQQQQQPN QNMSNANVPS1020
1021 DGFSLSQSQS MNFNQQQQQQ AAAQQQQVQP NMRQRQTQAQ AAAAAAAAAA QAQAAANASG1080
1081 PNVPLMQQPQ VGVGVGVGVG VGVGVGNGGV VGGPGSGGPN NGAMNQMGGP MGGMPGMQMG1140
1141 GPMNPMQMNP NAAGPTAQQM MMGSGAGGPG QVPGPGQGPN PNQAKFLQQQ QMMRAQAMQQ1200
1201 QQQHMSGARP PPPEYNATKA QLMQAQMMQQ TVGGGGVGVG GVGVGVGVGG VGGANGGRFP1260
1261 NSAAQAAAMR RMTQQPIPPS GPMMRPQHAM YMQQHGGAGG GPRTGMGVPY GGGAGGPMGG1320
1321 PQQQQRPPNV QVTPDGMPMG SQQEWRHMMM TQQQTQMGFG GPGPGGPMRQ GPGGFNGGNF1380
1381 MPNGAPNGAA GSGPNAGGMM SGPNVPQMQL TPAQMQQQLM RQQQQQQQQQ QQHMGPGAAN1440
1441 NMQMQQLLQQ QQSGGGGNMM ASQMQMTSMH MTQTQQQITM QQQQQFVQST TTTTHQQQQM1500
1501 MQMGPGGGGG GGGPGSANNN NGGGGGGAAG GGNSASTIAS ASSISQTINS VVANSNDFGL1560
1561 EFLDNLPVDS NFSTQDLINS LDNDNFNLQD FNMP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
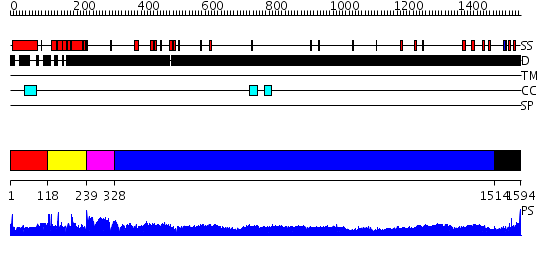
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..117] | 1.2 | crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution | |
| 2 | View Details | [118..238] | 5.154902 | No description for 2f8xM was found. | |
| 3 | View Details | [239..327] | 1.127996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [328..1513] | 95.69897 | No description for 1y0fA was found. | |
| 5 | View Details | [1514..1594] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.22 |
Source: Reynolds et al. (2008)