













| General Information: |
|
| Name(s) found: |
FBpp0086882
[FlyBase]
lat-PA / FBpp0086881 [FlyBase] |
| Description(s) found:
Found 34 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 721 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nuclear origin of replication recognition complex
[IDA][ISS]
|
| Biological Process: |
larval feeding behavior
[NAS]
DNA replication initiation [IDA] DNA replication [IEA][NAS] olfactory learning [TAS] learning [NAS] learning or memory [NAS] |
| Molecular Function: |
DNA binding
[IEA][NAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDPTISVSKG CFVYKNGATR AGKKAASKRK RPAAESSSLL GKEVVQQPFY EEYRKAWNQI 60
61 NDHIADLQHR SYARTLEQLV DFVVGQAERD TPDEVLPTAA LLTGINQPDH LSQFTALTQR 120
121 LHAQRAAMVC VLQSRDCATL KAAVETLVFG LVEDNAEVEQ MEDEDEDEDG AERDRKRLRR 180
181 SQCTMKQLKS WYTNNFDSEQ KRRQLVVILP DFECFNASVL QDLILILSAH CGSLPFVLVL 240
241 GVATAMTAVH GTLPYHVSSK IRLRVFQTQA APTGLNEVLD KVLLSPKYAF HLSGKTFKFL 300
301 THIFLYYDFS IHGFIQGFKY CLMEHFFGGN AFALCTDYSK ALGRIKQLTH EDMETIRRLP 360
361 SFRPYVEQIN DCKRIIAVLT DDDYLKKKLP QLLRDCLLHF LLFRCSLEFL TELVGDLPRC 420
421 PLGKLRRELY VNCLNRAIIS TPEYKECLQM LSFLSKDEFV AKVNRALERT EQFLVEEIAP 480
481 LELGEACTAV LRPKLEAIRL AVDEVVKATM ATITTTSPNE TRQATDHLTP VASRQELKDQ 540
541 LLQRSKEDKM RHQLNTPTTQ FGRALQKTLQ LIETQIVQDH LRALQDAPPI HELFVFSDIA 600
601 TVRRNIIGAP RAALHTALNN PHFYMQCKCC ELQDQSLLVG TLPDLSVVYK LHLECGRMIN 660
661 LFDWLQAFRS VVSDSDHEEV AQEQIDPQIQ ARFTRAVAEL QFLGYIKMSK RKTDHATRLT 720
721 W |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
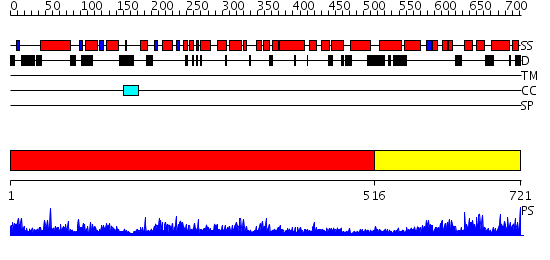
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..515] | 1.02 | CDC6, C-terminal domain; CDC6, N-domain | |
| 2 | View Details | [516..721] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.47 |
Source: Reynolds et al. (2008)