













| General Information: |
|
| Name(s) found: |
Caf1-105-PA /
FBpp0087402
[FlyBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 747 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chromatin assembly complex
[ISS][NAS]
|
| Biological Process: |
chromatin assembly or disassembly
[ISS]
nucleosome assembly [NAS] |
| Molecular Function: |
DNA binding
[NAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKCKIPEISW HNRDPVLSVD IQQNGLGLRS PTICRLASGG SDAHVLIWYV NRSDDAEGVD 60
61 VELAADLSRH QRAVNAVRWS PNGELLASGD DESVVFIWKQ KADHEVVNIV DADGCSEQDK 120
121 EVWLTLKVLR GHREDIYDLS WAPNSQFLVS GSVDNTAMLW DVHSGKSLAI LDDHKGYVQG 180
181 VAWDPCNQYI ATMSTDRQMR IFDANTKRVL HRVSKCVLPV KEDHEMHGKS MRLYQDGTLQ 240
241 TFFRRLCFTP DGKLLLTPSG ITDYDGVVKP INTSYGFSRH DLSKPAFVLP FPKEYAVAVR 300
301 CSPVLYRLRP YNAEKNPPII SLPYRMIYAV ATKNAVFFYD TQQPVPFAIV SNIHYSRLTD 360
361 LAWSSDGTVL IVSSTDGYCS LITFEPTELG DCYEDMETVL SVVLKSSENA TVLKKKRQKL 420
421 RKVSLDEPRK PLQEKSKPNT IRRASEAGIT EVGEPELDAE NDSSTHSVSS NKTNSPKTKA 480
481 SEEEKPTPIA IRRSPRKNST APMPIAIRRA PRKPEDKGNS ESSKPKDEME VTHTKTTQVQ 540
541 KTVASPVKRK VSTEAVPPAE TSQPALAVIP VFEKEIISSD DKFESPEKKS RPATPIQVRR 600
601 QPRTPGSSSQ FNLTPKSQPA KQATPIAVRR TPRVLIEMPV NSPVVPMEEA MDAWPLDESI 660
661 TPLPATKKDS RKPLPETVKT ETKPAVIEAS EATCERTEDI RLVYEDTQEE TPKNPTESGP 720
721 STPNNKTPRR VSLRTISTPK SKKKLLE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
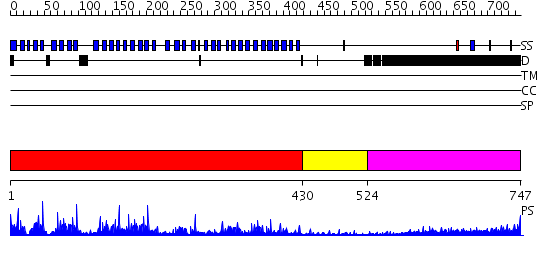
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..429] | 72.39794 | PAF-AH Holoenzyme: Lis1/Alfa2 | |
| 2 | View Details | [430..523] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [524..747] | 8.130997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)