













| General Information: |
|
| Name(s) found: |
KIME_RAT
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Rattus norvegicus |
| Length: | 395 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisome
[IDA]
soluble fraction [IDA] cytoplasm [IEA] cytosol [IDA] |
| Biological Process: |
regulation of cholesterol biosynthetic process
[NAS]
isopentenyl diphosphate biosynthetic process, mevalonate pathway [IDA] metabolic process [IEA] cholesterol biosynthetic process [TAS] isoprenoid biosynthetic process [UNKNOWN][IDA] phosphorylation [IEA] sterol metabolic process [TAS] negative regulation of translation [IDA] isoprenoid metabolic process [TAS] |
| Molecular Function: |
nucleotide binding
[IEA]
ATP binding [IDA] identical protein binding [UNKNOWN] mRNA binding [IDA] mevalonate kinase activity [UNKNOWN][IDA] transferase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSEVLLVSA PGKVILHGEH AVVHGKVALA VALNLRTFLV LRPQSNGKVS LNLPNVGIKQ 60
61 VWDVATLQLL DTGFLEQGDV PAPTLEQLEK LKKVAGLPRD CVGNEGLSLL AFLYLYLAIC 120
121 RKQRTLPSLD IMVWSELPPG AGLGSSAAYS VCVAAALLTA CEEVTNPLKD RGSIGSWPEE 180
181 DLKSINKWAY EGERVIHGNP SGVDNSVSTW GGALRYQQGK MSSLKRLPAL QILLTNTKVP 240
241 RSTKALVAGV RSRLIKFPEI MAPLLTSIDA ISLECERVLG EMAAAPVPEQ YLVLEELMDM 300
301 NQHHLNALGV GHASLDQLCQ VTAAHGLHSK LTGAGGGGCG ITLLKPGLER AKVEAAKQAL 360
361 TGCGFDCWET SIGAPGVSMH SATSIEDPVR QALGL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
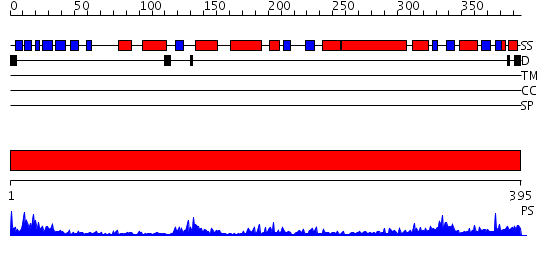
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..395] | 71.39794 | Mevalonate kinase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.69 |
Source: Reynolds et al. (2008)