













| General Information: |
|
| Name(s) found: |
MYF6_MOUSE
[Swiss-Prot]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Mus musculus |
| Length: | 242 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[TAS]
|
| Biological Process: |
muscle organ development
[IEA]
multicellular organismal development [IEA] somitogenesis [IMP] negative regulation of transcription [IMP] skeletal muscle tissue development [IMP][TAS] cell differentiation [IEA] regulation of transcription, DNA-dependent [IEA] positive regulation of transcription [IMP] regulation of transcription [IEA] skeletal muscle tissue regeneration [UNKNOWN] |
| Molecular Function: |
DNA binding
[IEA]
transcription regulator activity [IEA] RNA polymerase II transcription factor activity, enhancer binding [TAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMMDLFETGS YFFYLDGENV TLQPLEVAEG SPLYPGSDGT LSPCQDQMPQ EAGSDSSGEE 60
61 HVLAPPGLQP PHCPGQCLIW ACKTCKRKSA PTDRRKAATL RERRRLKKIN EAFEALKRRT 120
121 VANPNQRLPK VEILRSAISY IERLQDLLHR LDQQEKMQEL GVDPYSYKPK QEILEGADFL 180
181 RTCSPQWPSV SDHSRGLVIT AKEGGANVDA SASSSLQRLS SIVDSISSEE RKLPSVEEVV 240
241 EK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
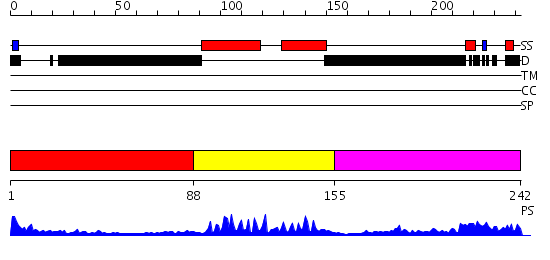
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..87] | 35.657577 | No description for PF01586.7 was found. No confident structure predictions are available. | |
| 2 | View Details | [88..154] | 15.39794 | Myod B/HLH domain | |
| 3 | View Details | [155..242] | 1.679997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)